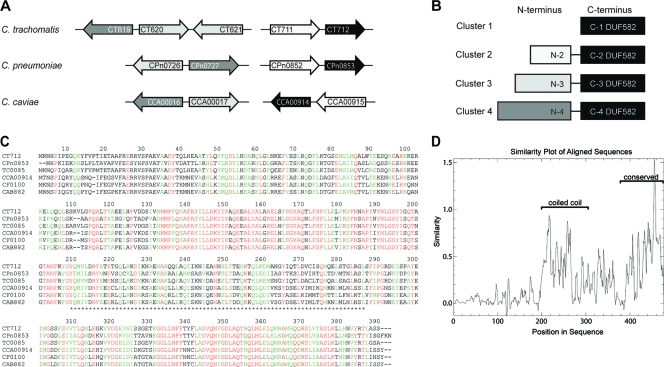
FIG. 1.
Conservation of the DUF582 proteins in Chlamydiaceae. (A) Schematic overview of the genes encoding DUF582-containing proteins in the C. trachomatis, C. pneumoniae, and C. caviae genomes. (B) Schematic representation of the Chlamydia DUF582 proteins. The proteins are grouped in four clusters based on sequence similarity. In all of them, DUF582 is located at the carboxy terminus. Proteins belonging to clusters 2, 3, and 4 contain additional N-terminal domains. The gray-level code corresponds to the DUF582 genes depicted in panel A. (C) Alignment of cluster 1 DUF582 proteins in six chlamydial genomes: C. trachomatis (CT), C. pneumoniae (CPn), C. muridarum (TC), C. caviae (CCA), C. felis (CF), and C. abortus (CAB). Identical residues are shown in red. Amino acids with high similarity are highlighted in green. Asterisks delimit the segment predicted to adopt a coiled-coil conformation. (D) Sequence conservation in the multiple alignment of all DUF582 sequences.