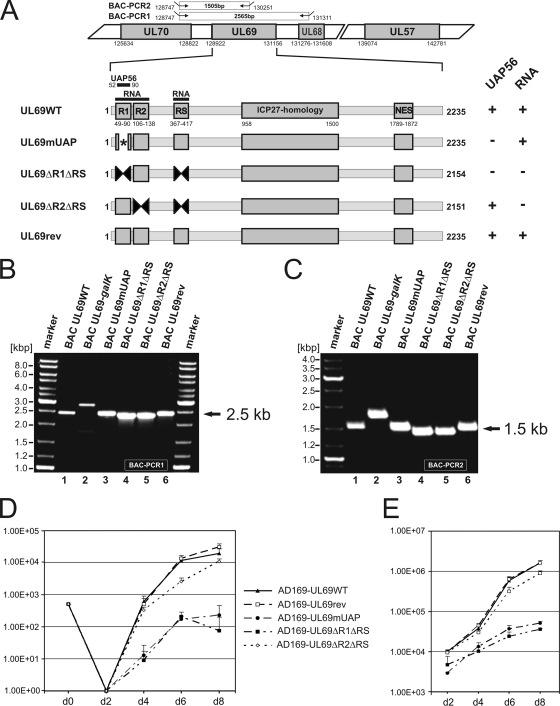
FIG. 9.
Growth kinetics of recombinant AD169-derived viruses carrying UL69 mutations that either affect the UAP56-binding capacity and/or the RNA-binding capacity of pUL69. (A to C) Construction and verification of recombinant HCMVs expressing either mutant or wild-type pUL69. (A) Schematic diagram illustrating the structure of recombinant viruses that were generated by homologous recombination in Escherichia coli. In a first step, the selection marker galK was inserted into UL69 of BAC pHB5. Subsequently, the selection marker was replaced by either a wild-type or a mutated UL69 sequence. The upper half of panel A shows the genomic region of HCMV strain AD169 containing UL69 (numbers refer to nucleotide positions of AD169; BAC-PCR1 and BAC-PCR2 refer to PCR amplifications that were performed to confirm the integrity of recombinant viruses). The lower half of panel A illustrates functional domains of UL69 and indicates the localization of mutations/deletions that were introduced into recombinant viruses (numbers refer to nucleotide positions of the UL69 open reading frame). UAP56, UAP56-binding motif of UL69; RNA, RNA-binding domain of UL69, comprising the arginine-rich motifs R1, R2, and RS; ICP27 homology, ICP27 homology region of UL69; NES, nuclear export signal. On the right of panel A, the UAP56- and/or RNA-binding capacities of pUL69 variants are indicated. (B and C) For verification of the correct recombination sites within the HCMV genome, PCR analyses of bacterial clones harboring the indicated BACs were performed using oligonucleotides specific for UL70/UL68 (B) or UL70/UL69 (C), respectively. The localizations of primers used for amplification are shown in the upper half of panel A (BAC-PCR1 and BAC-PCR2). (D) Growth kinetics of recombinant-AD169-derived viruses. In order to determine the replication capacities of the recombinant viruses AD169-UL69WT (wild-type), AD169-UL69rev (revertant), AD169-UL69mUAP (alanine substitution mutant of UAP56-binding motif), AD169-UL69ΔR1ΔRS (deletion mutant of arginine-rich motifs R1 and RS), and AD169-UL69ΔR2ΔRS (deletion mutant of arginine-rich motifs R2 and RS), multistep growth curve analyses were performed. HFFs were infected with equal IE units (MOI, 0.1) of wild-type, revertant, or mutant viruses. The viral supernatants were harvested at the indicated time points (days [d]), followed by the determination of viral titers. Each infection was performed in triplicate, and the standard deviations are depicted by error bars. (E) Quantification of viral genomes in the supernatants of infected HFF cells by real-time PCR. Aliquots of the supernatants obtained for the multistep growth curves shown in panel D were treated with proteinase K, incubated at 56°C for 1 h, and subsequently denatured at 95°C. Five microliters of each lysate was subjected to real-time PCR to quantify the genomic equivalents in the supernatants of the recombinant viruses. Evaluations were performed in triplicate for each of the three infections per virus. Standard deviations are indicated.