Fig. 3.
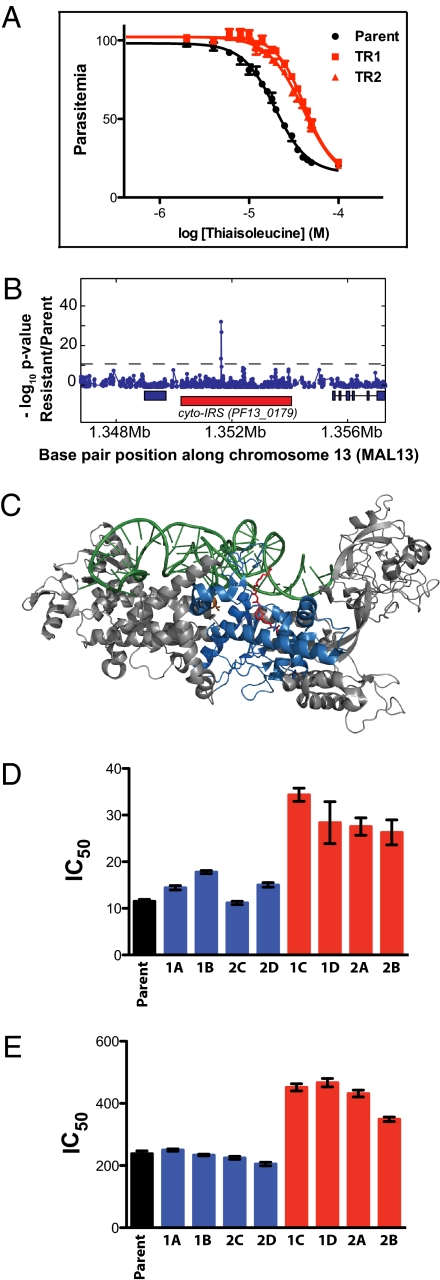
Thiaisoleucine-resistant parasites contain a mutation in the cyto-IRS gene, and the mutation is responsible for the resistance. (A) Sensitivity to thiaisoleucine of the 3D7 parent and two resistant clones (TR1 and TR2). Resistant parasites were selected in 20 μM thiaisoleucine/20 μM isoleucine. Clones were then grown in 20 μM isoleucine with varying concentrations of thiaisoleucine and parasitemia was normalized to a no inhibitor control. Data points of triplicates are shown as mean ± range. IC50 values and SEs are provided in SI Appendix, Table S3. (B) z test analysis of microarray. Genomic DNA of clone TR1 was digested with DNaseI and hybridized to a 25-mer oligonucleotide tiling microarray. Plot shows −log P value for z test (blue line) performed with TR1 versus 3D7 reference. The red rectangle below the z test indicates the position of PF13_0179; blue rectangles indicate neighboring genes. A single SNP in PF13_0179 (the gene encoding cyto-IRS) is detected. The A2430T mutation results in a L810F change. (C) Model of IRS from S. aureus [pdb: 1FFY (24)]. The active site is marked by bound mupirocin in red, tRNA in green, and conserved residues of the Rossmann fold in blue. The residue mutated in thiaisoleucine-resistant parasites (L810F) corresponds to a valine residue in the bacterial protein (V622; shown in orange). This model was generated with MacPyMOL (DeLano Scientific LLC). Note that cyto-IRS is only distantly related to the structure shown and that its closer homologs do not bind mupirocin. (D) and (E) Sensitivity of WT and 8 allelic replacement clones toward thiaisoleucine in 20 μM (D) or 180 μM isoleucine (E). Clones expressing WT cyto-IRS are shown in blue (1A, 1B, 2C, and 2D); clones expressing L810F cyto-IRS are shown in red (1C, 1D, 2A, and 2B). Error bars indicate SEs. Numeric values are given in SI Appendix, Table S4.