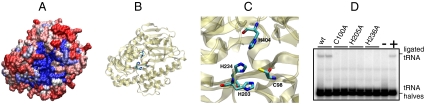
Fig. 4.
Crystal structure of P. horikoshii RtcB. (A) Surface model of the P. horikoshii RtcB crystal structure (taken from ref. 14). Sequence alignment of more than 50 archaeal and 40 eukaryotic RtcB proteins was used for plotting the sequence conservation from high (blue) to low (red) in the surface model. (B) The proposed active side residues forming a Zn2+ binding motif (12) are highlighted in the cartoon representation. (C) Closeup display of the putative Zn2+ coordinating cleft with the putative Zn2+-ligand amino acids (C98, H203, H234, H404). (D) tRNA ligase-activity assay of P. aerophilum wild-type and mutant (C100A, H205A, H236A; these residues correspond to C98, H203, H234 in the P. horikoshii sequence) MBP-RtcB enzymes. The tRNA splicing intermediates were incubated with the indicated P. aerophilum RtcB enzymes, without RtcB (lane −) or with the combined action of T4 polynucleotide kinase/3′-phosphatase and T4 RNA ligase 1 (lane +). The reaction products were analyzed by denaturating PAGE and subsequent PhosphorImager visualization.