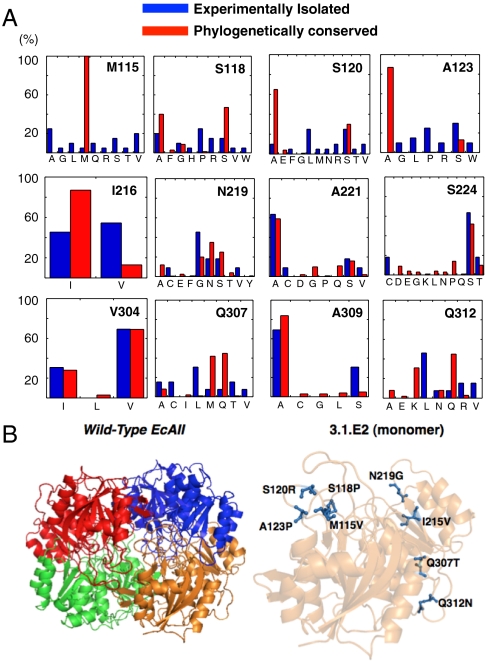
Fig. 2.
Isolation of EcAII neutral drift mutants. (A) Residue plasticity of known bacterial type II asparaginases (IPR004550; n = 478) and of EcAII variants exhibiting WT activity isolated by neutral drift (n = 20 per library) at the 12 amino acids targeted for mutagenesis. Residue numbers correspond to positions in EcAII. (B) Location of the eight amino acid mutations differentiating wild-type EcAII and 3.1.E2. Images generated by PyMol (55). The location of these mutations in relation to the EcAII active site is shown in Fig. S5.