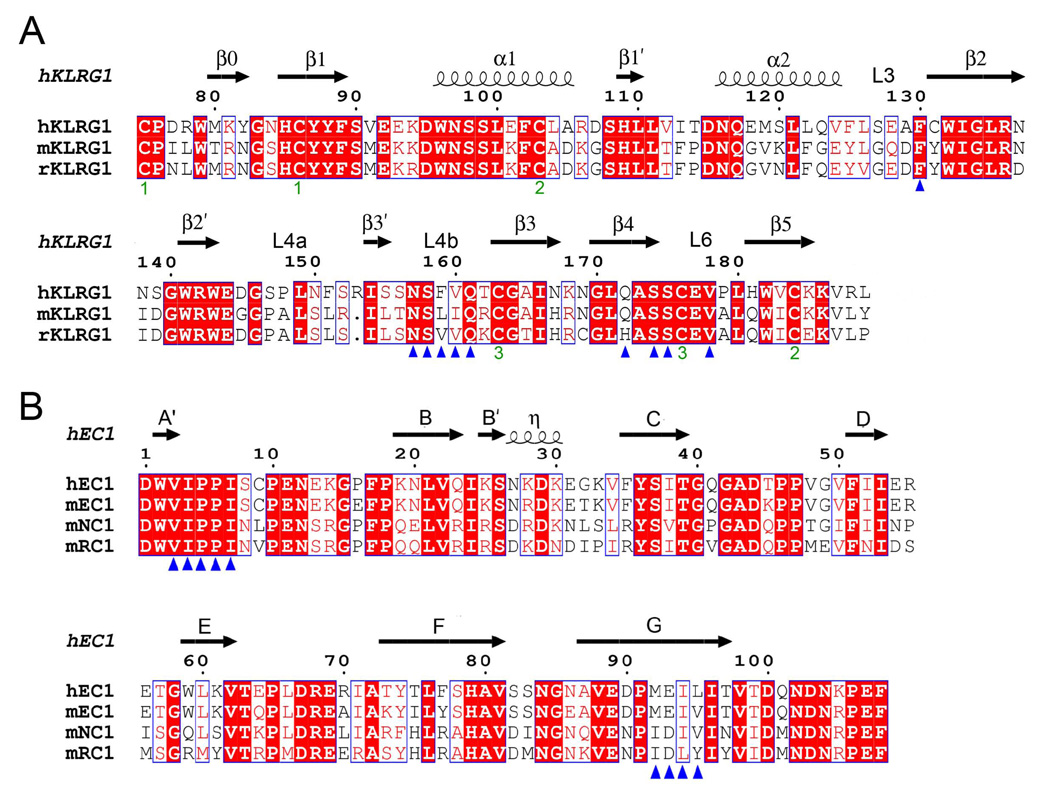
Figure 5. Structure-Based Sequence Alignments of KLRG1 Receptors and Cadherins.
(A) Sequence alignment of KLRG1 CTLDs. Secondary structure elements for hKLRG1 are denoted by squiggles (α-helices) and arrows (β-strands). These, and the loop regions, are numbered according to Figure 2A. Residues that contact E-cadherin in the mKLRG1–hEC1 and hKLRG1–hEC1 complexes are marked with blue triangles. The paired green numbers (1–3) indicate the bonded cysteine residues in the hKLRG1 and mKLRG1 structures. White characters on a red background show strictly residues. Residues that are well conserved are drawn in red and framed in blue. The remaining residues are black.
(B) Sequence alignment of the N-terminal domain of classical cadherins. Secondary structure elements for hEC1 are indicated by squiggles (310 helix (η)) and arrows (β-strands). Strands are labeled according to Figure 3A. Residues that contact KLRG1 in the mKLRG1–hEC1 and hKLRG1–hEC1 complexes are denoted with blue triangles. Sequences were retrieved from SwissProt using the following numbers: hKLRG1, Q96E93; mKLRG1, O88713; rat KLRG1 (rKLRG1) Q64335; hEC1, P12830; mEC1, P09830; mouse NC1 (mNC1), P15116; mouse RC1 (mRC1), P39038. Sequence alignments were performed with the program ClustalW (http://www.expasy.ch).