Figure 1.
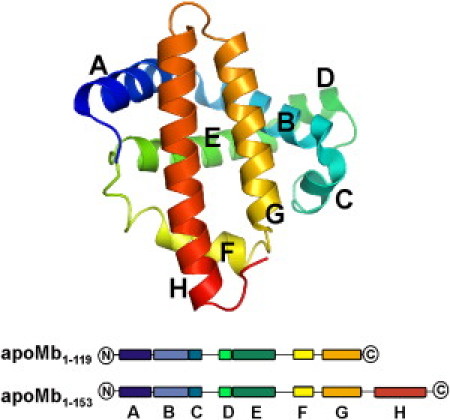
Three dimensional structure of apoMb. Atomic coordinates were derived from PDB file 1VXF, corresponding to the high-resolution crystal structure of carbonmonoxy-myoglobin (70). The image was created with the software PyMOL (DeLano Scientific, Palo Alto, CA). Below, a cartoon representation of the N-terminal fragment apoMb119 and full-length apoMb are given. The helices are color-coded and labeled from A to H (N- to C-terminus).
