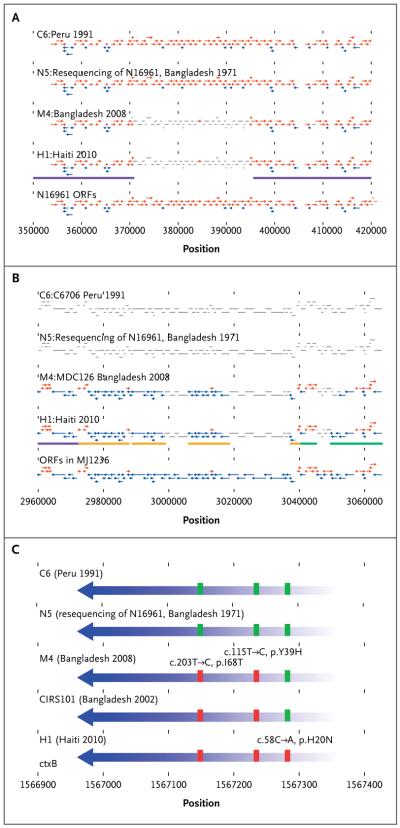
Figure 3. Gene Maps of the Superintegron, SXT, and ctxB Regions in the C6, N5, M4, and H1 V. cholerae Strains.
Representations of the coverage with respect to open reading frames (ORFs) in each of these regions are shown. Open reading frames that are shown in red are mapped to the positive strand, and those shown in blue are mapped to the negative strand; gray indicates that the open reading frame was supported by no reads or by a very low number of reads for the indicated sample. Coverages for H1 and H2 were identical over all regions; therefore, only H1 is shown in the figure. Colored bars below the H1 plots indicate the CIRS101 genomic region to which the reference sequence for the indicated element maps. Gaps indicate segments in the reference that are missing from CIRS101. For the SI region (Panel A), no coverage gaps were observed between the N16961 and the C6 and N5 strains, whereas a single coverage gap was observed in M4 and H1 from base positions 370682 to 395783 covering 41 open reading frames. For SXT (Panel B), none of the reads from N5 or C6 mapped to the MJ-1236 SXT reference sequence; in the case of H1, four coverage gaps were observed, covering 27 open reading frames. M4 had three gaps overlapping with three of the H1 gaps, but the M4 gaps covered 4 open reading frames in addition to 25 of the 27 open reading frames covered by the H1 gaps. Also shown (Panel C) is the location of all variant calls found in the cholera enterotoxin subunit B (VC_1456) open reading frame. The boxes indicate three sites in which the alleles represented in H1, CIRS101, and M4 (red) differ from the alleles in the N16961 and C6 sequences (green) and lead to nonsynonymous changes. In Panel C, c denotes the nucleotide position, and p the amino acid position.