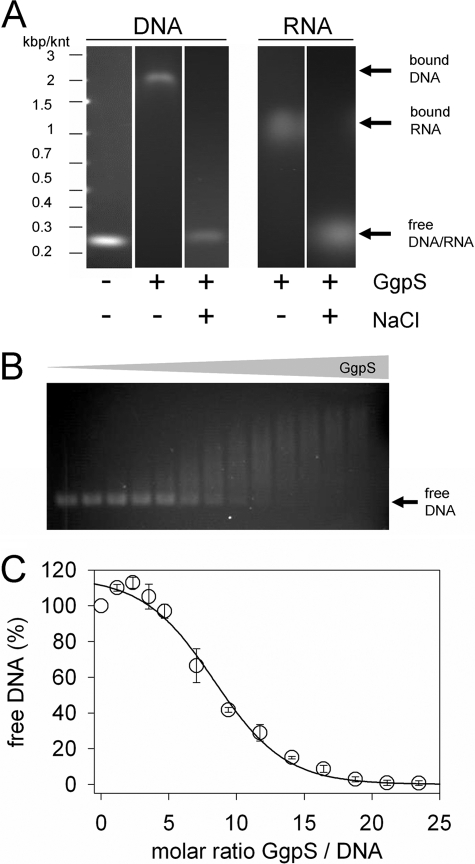
FIGURE 5.
Binding of GgpS to nucleic acids. EMSAs were performed using purified GgpS protein, a 233-bp DNA fragment obtained by PCR, or a 250-nucleotide RNA fragment obtained by in vitro transcription. Incubation and agarose gel electrophoresis were performed at low or high salt concentrations of 0 and 200 mm NaCl, respectively. GgpS (4.4 μm) and DNA (35 nm) or RNA (70 nm) were applied (A), or DNA (35 nm) was incubated with increasing GgpS concentrations (0–850 nm; B). The amount of free DNA at all of GgpS/DNA molar ratios was quantified by densitometry (C). The positions of shifted and nonshifted nucleic acids are indicated by arrows. kbp, kilobase pairs; knt, kilonucleotides.