FIGURE 6.
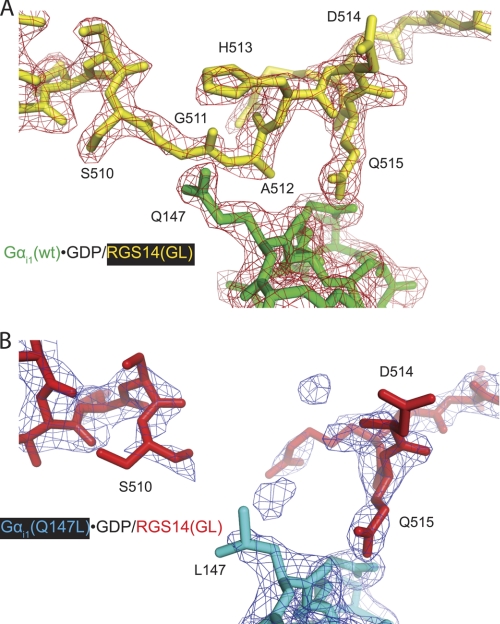
Differences in electron density observed within the GoLoco motif between WT and Q147L mutant Gαi1·GDP/RGS14 GoLoco motif complexes. A crystal structure (using diffraction data to 2.38-Å resolution) of the Gαi1Q147L·GDP/RGS14 GoLoco motif complex (Protein Data Bank code 3ONW) was obtained. Overall, the Gαi1Q147L protein binds the RGS14 GoLoco motif protein similarly (root mean square deviation of 0.38 Å) to Gαi1WT (code 2OM2 (7)). However, electron density for the GoLoco motif peptide residues Gly-511, Ala-512, and His-513 is much weaker in the Gαi1Q147L structure (B) than in the WT structure (code 2OM2) (A), indicating disorder in the peptide surrounding the mutated Gαi1 residue Leu-147. Both 2Fo − Fc electron density maps shown are contoured at σ = 1.5.