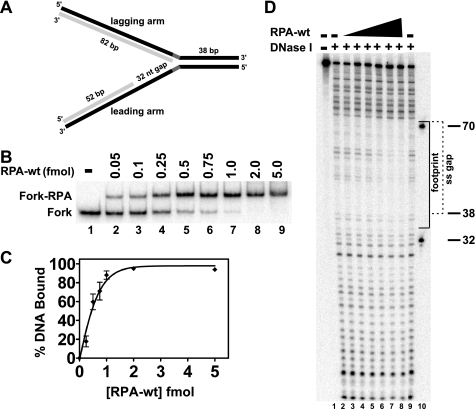
FIGURE 3.
RPA-wt binding to a replication fork substrate containing a single-stranded gap on the leading arm. A, the model replication fork substrate generated by two-step annealing of leadP122, lagP122, leadD52, and lagD82 oligonucleotides as indicated that contains a 32-nt single-stranded gap on the leading arm is shown, with parental and daughter strands depicted in black and light gray, respectively (for construction details and oligo sequences, see “Experimental Procedures” and supplemental Table 1, respectively). The parental-daughter arms were completely homologous except for a 5-nt non-complementary region (dark gray) at the fork junction to prevent spontaneous branch migration. The relevant lengths of the duplex and single-stranded regions are indicated. B, RPA-wt (0–5.0 fmol) was incubated with the fork substrate (1 fmol) for 10 min at 25 °C, and binding of RPA to the substrate was analyzed by EMSA as described under “Experimental Procedures.” The positions of the RPA-fork complexes and the fork substrate are indicated at the left. C, graphic representation of RPA binding to the fork substrate, derived from experiments performed as in B, calculated as the percentage of Fork-RPA complex compared with the total DNA for each reaction is shown. Data points are the mean of three independent experiments, except for values at 0.5 and 1 fmol of RPA-wt (four independent experiments). D, shown is RPA-wt (0.1–5 fmol) binding to the fork substrate (5 fmol) analyzed by DNase I footprinting as described under “Experimental Procedures.” The sizes of the markers (lane 10) and the boundaries of the leading arm gap (dashed bracket) and of the area of protection by RPA-wt (solid bracket) are denoted at the right.