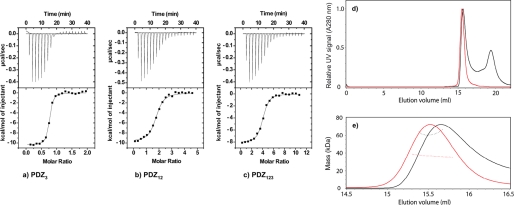
FIGURE 1.
Equilibrium binding experiments. a–c, ITC data for PDZ3 (a), PDZ12 (b), and PDZ123 (c) with E6L151V at 10 °C. Top panels, raw data. Bottom panels, integrated titration curves. See Table 1 for fitted parameters. d and e, SEC-MALS analysis of PDZ123 in the presence and absence of a 3-fold molar excess of HPV E6. d, the elution volumes from an analytical Superdex 75 10/30 column were very similar for the apo- and saturated PZD123-E6 complex (red and black traces, respectively). For clarity, the UV signals of PDZ123 peaks are normalized to have similar intensities. The additional peak at ∼19.4 ml corresponds to unbound HPV E6 (and reductants in the E6 samples used to prevent thiol-mediated oligomerization). e, SEC-MALS was used to determine the molar mass across the peaks for the apo- and saturated PZD123-E6 complex (trace colors are as in d). PDZ123 exists as a discrete monomer, even at high protein concentrations (>8 mg/ml). Despite having a similar elution volume to apo-PDZ123, the saturated PDZ123-E6 complex has a significantly increased molar mass. This mass difference was consistent with each PDZ123 binding three E6 molecules, which agrees with the number of binding sites available and independent ITC and kinetic measurements (Table 1).