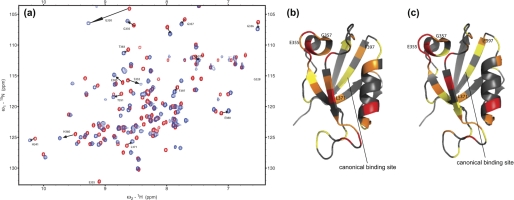
FIGURE 3.
NMR data of the E6-PDZ interaction. a, an overlay of HSQC spectra of free PDZ2 (red) and E6L151V-PDZ2 complex (blue). The arrows indicate residues that moved substantially and are discussed in the text. b and c, residues in PDZ2 undergoing structural change upon binding to the E6 protein. Color codes for chemical shift changes of residues in PDZ2 (Protein Data Bank code 2IOL) are: red, large change, Δ ≥ 0.15 ppm; orange, medium change, 0.10 ≤ Δ < 0.15 ppm; yellow, small change, 0.05 ≤ Δ < 0.10 ppm; and gray, no change, 0.00 ≤ Δ < 0.05 ppm. The PDZ2 domain was titrated with E6WT (b) and E6L151V (c). The four residues that display distinct chemical shift changes as compared with binding of a C-terminal peptide (33) are labeled. The canonical binding pocket is indicated by the solid line. This picture was drawn with PyMol (32).