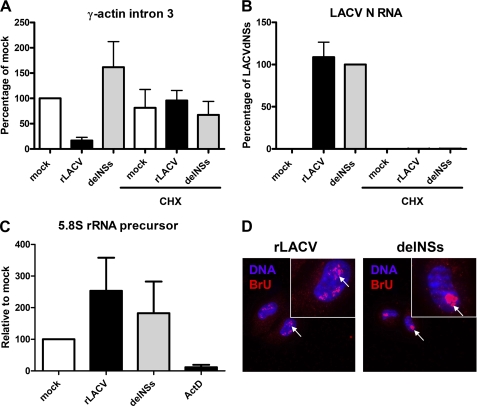
FIGURE 2.
NSs specifically suppresses RNAP II. A–C, real-time RT-PCR analysis is shown. 293T cells were seeded in 12-well plates to 80% confluency. At 1 h before infection, cells were either pretreated with 50 μg/ml cycloheximide (CHX) or left untreated. Then cells were either infected with rLACV or rLACVdelNSs (m.o.i. = 5) or mock-infected or treated with 1 μg/ml actinomycin D (ActD). After 4 h of incubation, total cell RNA was extracted, and real-time RT-PCR was performed using primers specific for γ-actin intron 3 (A), LACV N RNA (B), or the 5.8 S rRNA precursor (C). All values obtained were normalized against γ-actin mRNA levels. Up-regulation of inducible genes is depicted in relation to mock-infected cells. Mean values and S.D. from three independent experiments are shown. D, an in situ run-on assay is shown. HeLa cells were grown on coverslips and infected with rLACV or rLACVdelNSs (m.o.i. = 1) for 8 h. After permeabilization with buffer containing Triton X-100, cells were incubated with buffer containing Br-UTP to label newly synthesized RNA and α-amanitin to quench RNAP II. After 30 min of labeling, cells were fixed and analyzed by immunofluorescence using an antibody against BrU (red signal) and counterstained with DAPI (blue signal). Insets show a magnification of individual nuclei, and arrows indicate the nucleoli containing de novo synthesized RNAP I transcripts.