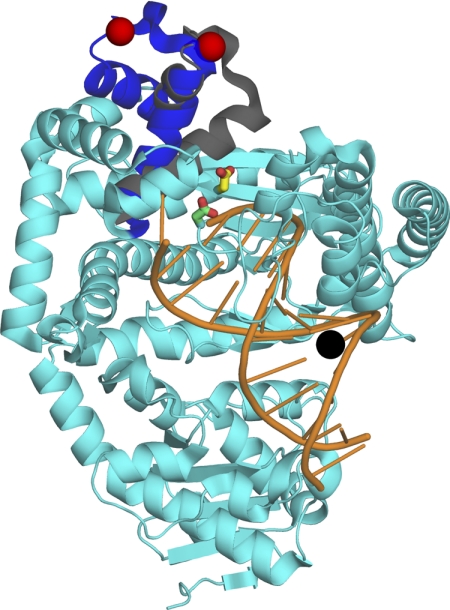
FIGURE 2.
Experimental design and the position of the mutants. The α-carbons of the binary complex (Protein Data Bank entry 1L3U) and of the ternary complex (Protein Data Bank entry 1LV5) of Bst DNA polymerase (6) were aligned using PyMOL (DeLano Scientific LLC). Both complexes were essentially identical except for the mobile portion of the fingers subdomain (residues 680–714, equivalent to 732–766 of Pol I(KF), shown in blue in the binary and gray in the ternary complex; the non-mobile portion of the protein (from 1L3U) is shown in cyan. The residues Asp705 and Asp882 of Pol I(KF), used in our study, correspond to the residues Asp653 and Asp830 of Bst pol, colored in yellow and green, respectively. The red spheres indicate the attachment point for the donor fluorophore on the protein (residue 692 of Bst pol, equivalent to residue 744 of Pol I(KF)). The DNA primer-template is shown in orange, with the attachment position of the dabcyl quencher on the template strand shown as a black sphere.