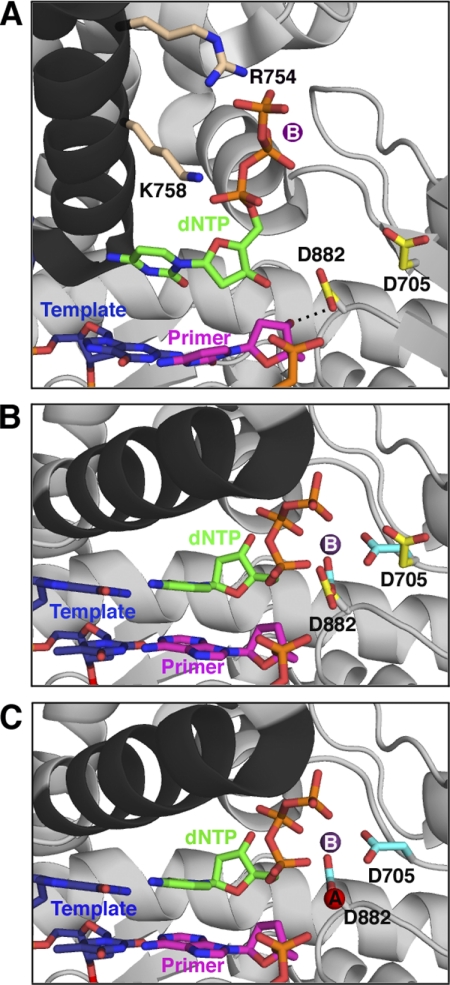
FIGURE 8.
Proposed sequence of events for the prechemistry steps involving Asp882 and Asp705. All protein structures are based on crystal structures of Bst pol, but side chains are labeled with Pol I(KF) residue numbers; the protein backbone is shown in gray, except for the O-helix (black). A, the initial, open, ternary complex was modeled from the binary complex (Protein Data Bank entry 1L5U) (6) with the position of the nucleotide (green) taken from the structure of Klentaq bound to dCTP in the absence of DNA (Protein Data Bank entry 5KTQ) (19). The nucleotide's triphosphate tail is bound by the conserved side chains Arg754 and Lys758 (beige) on the O-helix. The Asp882 and Asp705 side chains are shown in yellow; Asp882 is hydrogen-bonded (dashed line) to the 3′-OH of the primer strand, whereas Asp705 points away from the active site. The primer strand of the DNA is shown in pink, and the template strand is shown in blue. A suggested position for metal B (purple), chelated via the dNTP β- and γ-phosphates, is indicated (no bound metal is present in the 5KTQ coordinates). B, the ternary complex (taken from Protein Data Bank entry 1LV5) (6) immediately after the fingers have closed but before metal A binds, showing the change in position of the Asp705 and Asp882 side chains, upon going from the open (yellow, as in A) to closed (cyan) conformation. Arg754 and Lys758 remain in contact with the incoming nucleotide in the closed conformation but are omitted for clarity. Since the primer is dideoxy-terminated in this structure, the interaction between the 3′-OH group and the carboxylate of Asp882 is absent. C, the closed complex is illustrated (as in B), showing the entry of metal A (red, from 1LV5). This structure represents the enzyme poised for catalysis.