FIG. 4.—
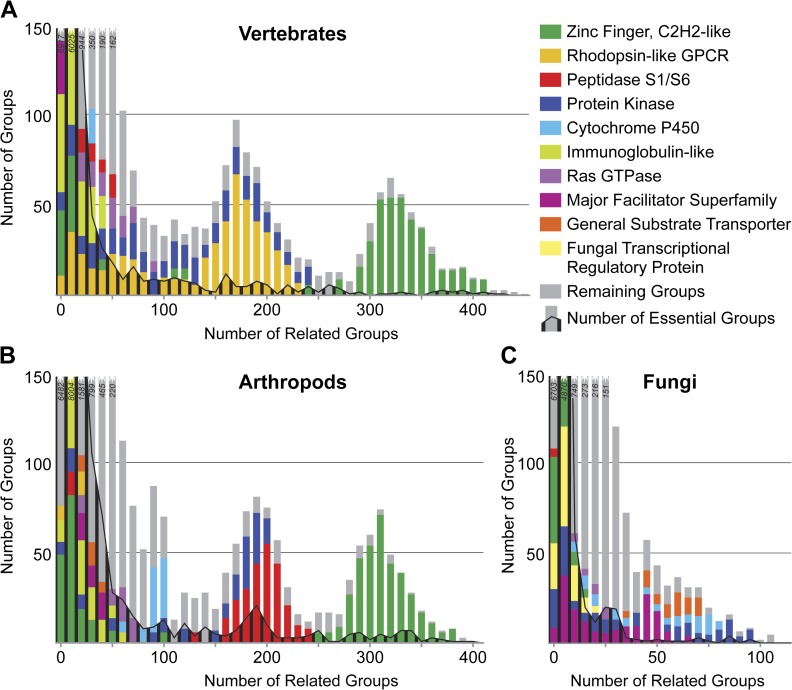
Independent proliferation of a few functional domains has created distinguishing protein superfamilies in vertebrates (A), arthropods (B), and fungi (C). The majority of the orthologous groups with identifiable homology to other groups are related to only a few other groups, however, those with numerous relatives are often characterized by specific protein domains. The superfamily of zinc finger C2H2-like proteins is common to vertebrates and arthropods, whereas the expansion of vertebrate rhodopsin-like GPCRs contrasts that of the arthropod peptidases. The fungi are characterized by families of transporters of the major facilitator superfamily and general substrate transporters. Cytochrome P450s are prominent in arthropods and fungi and the protein kinases feature in all three lineages. Orthologous groups with essential genes may be found among those with some of the highest numbers of relatives. Related groups are defined by the average pairwise Smith–Waterman e value between all the members of each group in each lineage with a cutoff of 1 × 10−3.