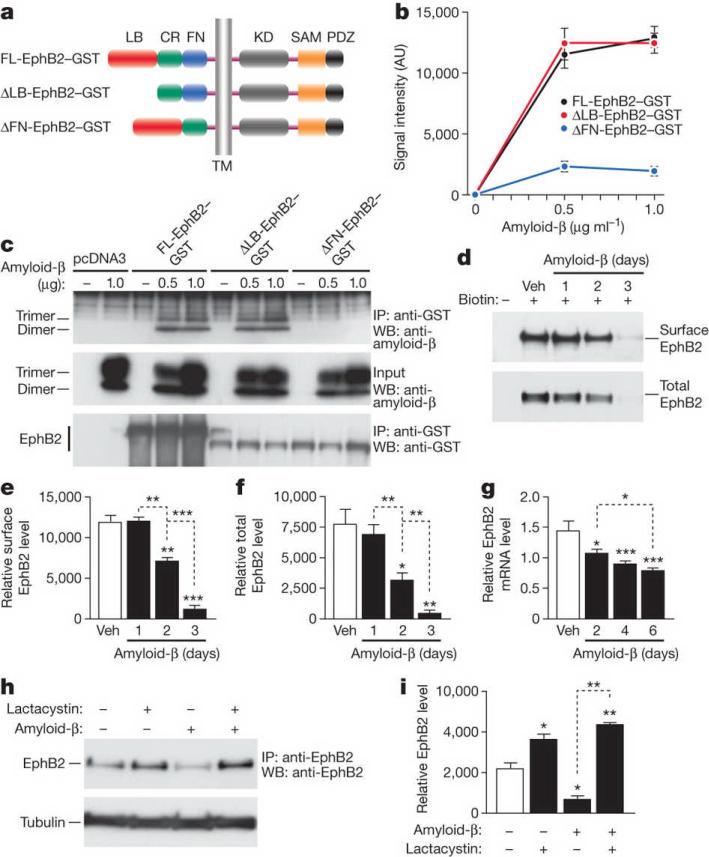
Figure 1. Amyloid-β oligomers bind to the fibronectin repeats domain of EphB2 and cause degradation of EphB2 in the proteasome.
a, Domain structure of full-length (FL) EphB2 and deletion constructs. Ligand-binding (LB) domain, cysteine-rich (CR) region, fibronectin type III repeats (FN) domain, transmembrane (TM) region, tyrosine kinase (KD) domain, sterile alpha motif (SAM) domain, and PSD95, DLG and ZO1 (PDZ) domain. b, Binding of amyloid-β dimers and trimers to different EphB2 constructs. See Supplementary Table 2 for experimental details pertaining to data shown in figures. AU, arbitrary units. c, Representative western blot (WB). IP, immunoprecipitation. d–f, Amyloid-β-induced depletion of EphB2. Primary rat neurons were treated with amyloid-β or vehicle (Veh), and surface and total levels of EphB2 were determined by western blots. Representative western blots are shown in d. e, f, Quantification of surface (e) and total (f) levels of EphB2. g, EphB2 mRNA levels in primary neurons treated with amyloid-β or vehicle. h, i, Lactacystin blocks amyloid-β-induced depletion of EphB2 in primary neurons. Representative western blot (h) and quantification of signals (i). For all experiments, n = 3–6 wells per condition from three independent experiments. *P < 0.05, **P < 0.001, ***P < 0.0001 versus empty bars or as indicated by brackets (Tukey test). Values are means ± s.e.m.