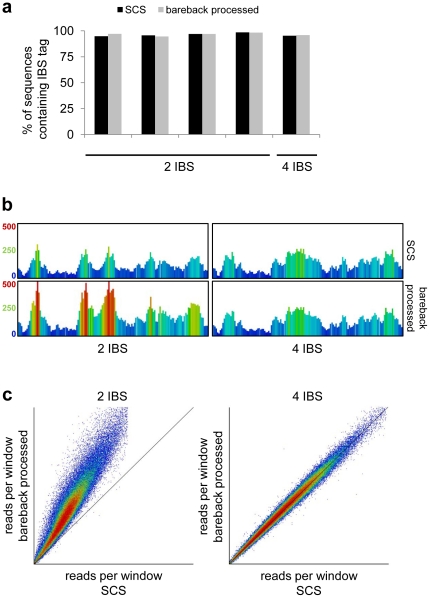
Figure 4. Bareback-processing increases sequencing depth without introducing bias.
(a) The percentage of sequences containing the expected initially biased sequence (IBS) identifiers, processed with either the standard SCS pipeline or subjected to bareback-processing, shuffling the first four bp to the end of the sequence. (b) Comparison of the sequencing depth in a 14 kb region for two- and four-IBS library samples after SCS- or bareback-processing, followed by sequence alignment. Each bar represents a window of 100 bp, and the heat map colours range between 20 and 500 sequencing reads. (c) Scatter plot representation comparing the read count distribution of SCS- vs. bareback-processed samples with either two or four different IBS. Reads of an entire flow-cell lane were counted in sliding windows of 100 bp.