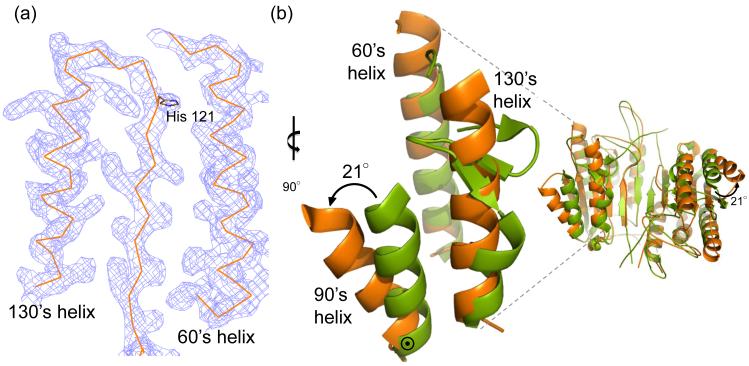
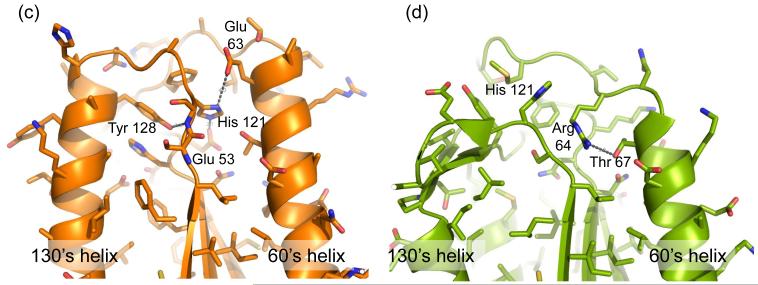
Fig. 4.
Unique structural features of caspase-6. (a) 2Fo-Fc electron density map contoured at 1 σ in the region of the 60’sand 130’s helices. Cα trace is shown in orange. (b) Comparison of mature ligand-free caspase-6 60’s, 90’s and 130’s helices (orange) to the homologous region of mature ligand-free caspase-7 (1K86, green). ⊙ denotes the hinge around which the 90’s helix pivots by 21°. (c) Interactions (dashes) holding the 60’s and 130’s network of helices together are mediated by the inactive conformation of catalytic-dyad residue, His 121. (d) The homologous region to (c) in the active-site liganded caspase-7 structure with caspase-6 numbering shown. Indicated residues (drawn in sticks) have the same amino acid identity in both caspase-6 and caspase-7, although the numbering is different. R64, T67 and H121 in caspase-6 numbering are R87, T90 and H144 in caspase-7 numbering. (e) Sequence alignment for the 60’s, 90’s and 130’s helices for all caspases. Amino acid numbering for each caspase is indicated. Strictly conserved residues (orange letters), important network residues (gray highlight) and the catalytic histidine (yellow highlight) in apoptotic initiators, executioners (Exec) and inflammatory (Inflam) caspases are shown. (f) Surface of caspase-6 near the 90’s helix (drawn with 2WDP coordinates, in which the 90’s helix side chains are better resolved) showing the pocket between the 90’s and 130’s helices