Telomeres are nucleo-protein structures that protect chromosome ends against degradation, fusion, recombination and recognition by the DNA damage machinery and at the same time facilitate replication of the ends of linear eukaryotic chromosomes.1 The maintenance of telomere length depends on telomerase, a ribonucleoprotein complex that uses its RNA template to elongate the telomere by addition of G-rich telomeric repeats to the terminal 3′ overhang. Synthesis of the complementary 5′ C-rich telomeric strand is driven by conventional DNA polymerases.2
In the budding yeast Saccharomyces cerevisiae, the ends of chromosomes contain a 250–300 bp array of TG1–3 repeats. The extreme ends of telomeres consist of a 12–14 nucleotides 3′ single-stranded overhang except in late S phase when longer overhangs are detected.3 The essential repressor/activator protein 1, Rap1, is specifically associated with telomeric duplex DNA repeats4 while the single-stranded G-rich overhang is bound by Cdc13 belonging to the CST complex (Cdc13/Stn1/Ten1) that plays a major role in telomere end protection and telomere elongation.5 The CST complex protects telomere ends and recruits DNA polymerase alpha, which synthesizes the complementary C-strand by virtue of the ability of Cdc13 to interact with the Stn1/Ten1 proteins. Cdc13 is also essential for telomere access by the telomerase complex through an interaction with Est1, a subunit of the telomerase holoenzyme that contains the protein catalytic subunit Est2 and its integral RNA template TLC1.6 In budding yeast, G-tail formation involves the MRX (Mre11/Rad50/Xrs2) complex, which controls one of the two distinct but partially complementary pathways of nucleolytic resection at telomeres.7 In addition, the MRX complex has been shown to be required for the robust association of telomerase with telomeres, by promoting the binding of the kinase Tel1 to short telomeres via a specific interaction with Xrs2.8 Binding of the MRX complex to telomeres has been shown to be negatively regulated by long telomeric tracts.9 Therefore, binding of MRX is favoured at short telomeres and is sufficient to explain the binding of Tel1 to these ends that are therefore preferentially elongated by telomerase.10
The notable difference between telomeres and DNA double-strand breaks (DSBs) is in the cellular response to these structures. Whereas telomeres are relatively inert and stable, DSBs are immediately recognized by the MRX complex, leading to Tel1 activation. In S/G2 phase, DSBs are extensively resected into 3′ single-stranded overhangs, which are coated by replication protein A (RPA). RPA is the lynchpin in subsequent cell cycle checkpoint and recombinational repair events by recruiting the Mec1-Ddc2 and Ddc1-Rad17-Mec3 checkpoint complexes, and the key recombination protein Rad52. In budding yeast, Rad52 is required for the recruitment of downstream recombination factors including the Rad51 recombinase, Rad59, the Rad55–Rad57 mediator and the Rad54 SWI/SNF homologue.11
In budding yeast lacking telomerase, telomeres reach a critical short length and enter replicative senescence after about 50 generations.12 Many factors cause telomere erosion. First, the lagging strand replication machinery is unable to fully copy the parental strand. Second, the parental strand replicated by the leading strand machinery is thought to be trimmed by a 5′ exonuclease activity directly coupled to passage of the replication fork. Third, the various chromatin structures that form at telomeres are a source of difficulties for passage of the replication fork.13 Several reports suggest that replication forks can pause naturally at telomeres in both budding and fission yeast.14–16 Current models stipulate that replicative senescence results from uncapped telomeres being recognized as DSBs. In a recent study, we have shown by single cell analysis and chromatin immunoprecipitation that loss of telomerase is accompanied by an ordered recruitment of Mre11 protein, Cdc13 the telomere sequence-specific single-stranded DNA binding protein, RPA, the ATRIP-like Ddc2 checkpoint protein, and the Rad52 recombination protein.17 We observed that this DNA damage response at telomeres starts many generations before the onset of senescence and that a single short telomere is sufficient to induce the recruitment of recombination proteins. Interestingly, a single Cdc13 focus is observed during the collective shortening of all telomeres in telomerase-negative cells without affecting the organization of telomeres into 4–6 clusters at the nuclear periphery. To explain these findings, we have proposed two models. Firstly, a burst of resection could occur at a single telomere in S phase among a population of cells exhibiting a gradual shortening of telomeres. The predominant binding of Cdc13 rather than RPA to the generated single-stranded TG1–3 overhangs likely prevents the formation of a recombination-proficient RPA filament and subsequent recruitment of the Rad52 recombination protein. Only when resection extends into subtelomeric regions, RPA and Rad52 are recruited to initiate homologous recombination. In a second model, the early telomere damage response could result from accidental replication fork collapse at a telomere, which would expose ssDNA being recognized by Cdc13 and RPA. In the absence of telomerase, recruitment of Rad52 would serve as an alternative pathway to repair telomeres after such accidental replication fork collapse.
The other striking observation of our study is that the eroded telomeres, while remaining at the nuclear periphery, move from their membrane anchor sites to the nuclear pore complexes (NPCs).17 This association is strongest after about 55 generations after the loss of TLC1 expression and ends approximatively after 70 generations, e.g., during the time by when survivors arise. In telomerase positive cells, budding yeast telomeres are normally clustered into 4–6 foci at the nuclear periphery through two redundant pathways involving the silent information regulatory protein Sir4 and the yKu heterodimer.18,19 The anchoring of telomeres to the nuclear envelope mediated by Sir4 relies on its interaction with Esc1, a protein exclusively associated with the inner face of the nuclear envelope19 while the perinuclear anchoring activity of yKu depends on the interaction between telomere-bound telomerase and the integral nuclear membrane protein Mps3.20 Interestingly, in an independent study, Oza et al. (2009) demonstrated that unrepaired or slowly repaired DSBs formed within an asynchronous cell population are sequestered at the nuclear envelope through a process that requires the telomerase holoenzyme and Mps3.21 Indeed, robust recruitment of Cdc13 and Est2 to the irreparable HO-induced DSB was shown with kinetics that follow the recruitment of Mps3. Furthermore, recruitment of Cdc13 to the DSB was shown to require the activity of Mre11 and the recombination protein Rad51 suggesting that in the absence of Rad51, RPA may prevent Cdc13 occupancy at the DSB. Another concomitant study reveals that at a persistent DNA break, after DNA resection and RPA recruitment, Rad51 spreads chromosome-wide bidirectionally from the DSB. The persistent DSB is later associated with the nuclear periphery in the vicinity of Mps3, in a process that requires Rad51 and the histone variant H2A.Z.22 H2A.Z is largely evicted from the break hours before the DSB relocalizes to the periphery suggesting that eviction of H2A.Z facilitates a signal transduction pathway that triggers relocalization to the nuclear envelope.22 These studies parallel those from S. Gasser's group showing that a unique irreparable HO-induced DSB or replication fork-associated breaks relocalize to the NPC. This localization depends on an intact NPC, the ATR-related kinase Mec1 and on the SUMO-regulated Slx5/8 ubiquitin ligase complex.23 Moreover, a previous study indicated that Mec1 accumulates to an irreparable HO-induced DSB via an interaction between its associated protein Ddc2 (ATRIP) and the single-strand binding protein RPA and that Rad51 reduces Mec1 accumulation at the processed break,24 indicating that Rad51 filaments and RPA coating represent competing structures at DSBs and potentially also at eroded telomeres.
In this context, we show that telomerase-negative cells bearing a single very short telomere senesce earlier, demonstrating that the length of the shortest telomere in the cell is a major determinant of the onset of senescence. Remarkably, we observed that some cells keep on dividing for some generations despite a complete erosion of their very short telomere. From these results, we have proposed that in the absence of telomerase, a very short telomere is first maintained in a pre-signaling state and then switches to a signaling state leading to senescence through a Mec1-dependent pathway.25
Collectively these papers suggest that unrepaired DSBs as well as eroded telomeres may exist in two different states leading to distinct fates. The first state would promote telomerase recruitment, Mps3-dependent tethering to the nuclear envelope and telomere addition, while the second state would lead to Mec1 recognition, checkpoint activation, and relocalization to the NPC, where recombinational repair is promoted (Fig. 1). Since the recruitment of Mec1 to irreparable breaks is mediated by RPA while telomerase recruitment depends on Cdc13 and yKu,26,27 we speculate that RPA, yKu and Cdc13 may compete for the binding to resected telomeres to direct their fate.
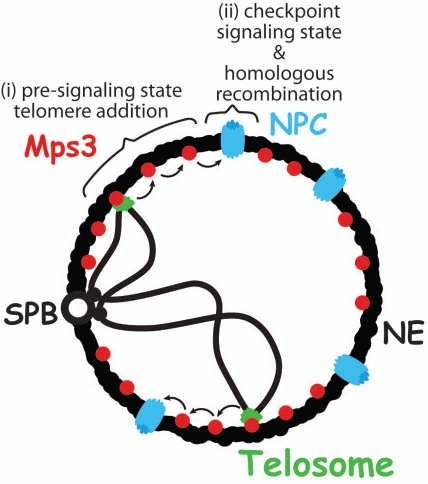
Figure 1.
Model for two-step response to eroded telomeres. Initially, eroded telomeres are maintained in a pre-signaling state via Mps3-dependent tethering at the nuclear envelope (NE). This state promotes telomere addition by telomerase (i). Subsequently, persisting eroded telomeres relocalize to the nuclear pore complex (NPC), where homologous recombination is promoted, and they switch to a Mec1-dependent signaling state, leading to replicative senescence (ii). SPB, spindle pole body.
The role of the Mps3-dependent tethering would be to protect chromosome ends from erroneous telomere recombination by stimulating telomerase activity, while relocalization to the NPC would favor recombination as a last resort instead of telomere fusion by non-homologous end-joining. Indeed, genetic studies indicate that the Rad52 recombination pathway becomes essential in yeast carrying nuclear pore deficiencies.28 Moreover, it has been shown that telomere tethering at the nuclear periphery was essential for efficient DSB repair in subtelomeric region29 and that nucleoporins are required for the establishment of appropriate sumoylation of several cellular proteins by maintaining proper levels of the SUMO-deconjugating enzyme Ulp1 at NPCs.30 Both increase and decrease in the levels of SUMO-conjugates were observed in NPC mutants as well as in a ulp1 mutant.30 Taken together these results indicate that the nuclear pore complex may play a role in recombinational repair of DSBs and eroded telomeres by modulating the sumoylation of DNA repair proteins. Interestingly, the potential role of homologous recombination in repairing these short telomeres is likely relevant also to wild-type cells, where colocalizing Cdc13 and Rad52 foci are frequently observed.17
One may raise the question whether relocalization of the persistent DSBs to the Mps3 sites at the nuclear envelope precedes association with the NPC as proposed by Gartenberg.31 In support of this model the results from Oza et al. (2009) suggest that a persistent DSB first experiences telomere addition, but if the end lacks sufficient TG repeats, it then hands the DSB off to the NPC where alternative repair pathways are likely to occur.21 Therefore irreparable DSBs and eroded telomeres appear to be channeled into the same pathway.
Acknowledgements
We thank members of the Géli, Gilson and Lisby laboratories for their input in this work. We also thank Mauro Modesti and Craig Peterson for ideas and discussions. This work was supported by The Danish Agency for Science, Technology and Innovation (M.L.), the Villum Kann Rasmussen Foundation (M.L.); V.G. and E.G. laboratories are supported by the “Agence Nationale de la Recherche”, (ANR programme blanc) and by the “Ligue Nationale contre le Cancer (LNCC), (équipes labelisées)”.
Footnotes
Previously published online: www.landesbioscience.com/journals/nucleus/article/11173
References
- 1.Longhese MP. DNA damage response at functional and dysfunctional telomeres. Genes Dev. 2008;22:125–140. doi: 10.1101/gad.1626908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McEachern MJ, Krauskopf A, Blackburn EH. Telomeres and their control. Annu Rev Genet. 2000;34:331–358. doi: 10.1146/annurev.genet.34.1.331. [DOI] [PubMed] [Google Scholar]
- 3.Larrivee M, LeBel C, Wellinger RJ. The generation of proper constitutive G-tails on yeast telomeres is dependent on the MRX complex. Genes Dev. 2004;18:1391–1396. doi: 10.1101/gad.1199404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Conrad MN, Wright JH, Wolf AJ, Zakian VA. RAP1 protein interacts with yeast telomeres in vivo: overproduction alters telomere structure and decreases chromosome stability. Cell. 1990;63:739–750. doi: 10.1016/0092-8674(90)90140-a. [DOI] [PubMed] [Google Scholar]
- 5.Linger BR, Price CM. Conservation of telomere protein complexes: shuffling through evolution. Crit Rev Biochem Mol Biol. 2009;44:434–446. doi: 10.3109/10409230903307329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bianchi A, Shore D. How telomerase reaches its end: mechanism of telomerase regulation by the telomeric complex. Mol Cell. 2008;31:153–165. doi: 10.1016/j.molcel.2008.06.013. [DOI] [PubMed] [Google Scholar]
- 7.Bonetti D, Martina M, Clerici M, Lucchini G, Longhese MP. Multiple pathways regulate 3′ overhang generation at S. cerevisiae telomeres. Mol Cell. 2009;35:70–81. doi: 10.1016/j.molcel.2009.05.015. [DOI] [PubMed] [Google Scholar]
- 8.Sabourin M, Zakian VA. ATM-like kinases and regulation of telomerase: lessons from yeast and mammals. Trends Cell Biol. 2008;18:337–346. doi: 10.1016/j.tcb.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Negrini S, Ribaud V, Bianchi A, Shore D. DNA breaks are masked by multiple Rap1 binding in yeast: implications for telomere capping and telomerase regulation. Genes Dev. 2007;21:292–302. doi: 10.1101/gad.400907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Teixeira MT, Arneric M, Sperisen P, Lingner J. Telomere length homeostasis is achieved via a switch between telomerase-extendible and -nonextendible states. Cell. 2004;117:323–335. doi: 10.1016/s0092-8674(04)00334-4. [DOI] [PubMed] [Google Scholar]
- 11.Lisby M, Rothstein R. Choreography of recombination proteins during the DNA damage response. DNA Repair (Amst) 2009;8:1068–1076. doi: 10.1016/j.dnarep.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.AS IJ, Greider CW. Short telomeres induce a DNA damage response in Saccharomyces cerevisiae. Mol Biol Cell. 2003;14:987–1001. doi: 10.1091/mbc.02-04-0057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gilson E, Geli V. How telomeres are replicated. Nat Rev Mol Cell Biol. 2007;8:825–838. doi: 10.1038/nrm2259. [DOI] [PubMed] [Google Scholar]
- 14.Makovets S, Herskowitz I, Blackburn EH. Anatomy and dynamics of DNA replication fork movement in yeast telomeric regions. Mol Cell Biol. 2004;24:4019–4031. doi: 10.1128/MCB.24.9.4019-4031.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller KM, Rog O, Cooper JP. Semi-conservative DNA replication through telomeres requires Taz1. Nature. 2006;440:824–828. doi: 10.1038/nature04638. [DOI] [PubMed] [Google Scholar]
- 16.Chang M, Arneric M, Lingner J. Telomerase repeat addition processivity is increased at critically short telomeres in a Tel1-dependent manner in Saccharomyces cerevisiae. Genes Dev. 2007;21:2485–2494. doi: 10.1101/gad.1588807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khadaroo B, Teixeira MT, Luciano P, Eckert-Boulet N, Germann SM, Simon MN, et al. The DNA damage response at eroded telomeres and tethering to the nuclear pore complex. Nat Cell Biol. 2009;11:980–987. doi: 10.1038/ncb1910. [DOI] [PubMed] [Google Scholar]
- 18.Hediger F, Neumann FR, Van Houwe G, Dubrana K, Gasser SM. Live imaging of telomeres: yKu and Sir proteins define redundant telomere-anchoring pathways in yeast. Curr Biol. 2002;12:2076–2089. doi: 10.1016/s0960-9822(02)01338-6. [DOI] [PubMed] [Google Scholar]
- 19.Taddei A, Hediger F, Neumann FR, Bauer C, Gasser SM. Separation of silencing from perinuclear anchoring functions in yeast Ku80, Sir4 and Esc1 proteins. EMBO J. 2004;23:1301–1312. doi: 10.1038/sj.emboj.7600144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schober H, Ferreira H, Kalck V, Gehlen LR, Gasser SM. Yeast telomerase and the SUN domain protein Mps3 anchor telomeres and repress subtelomeric recombination. Genes Dev. 2009;23:928–938. doi: 10.1101/gad.1787509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oza P, Jaspersen SL, Miele A, Dekker J, Peterson CL. Mechanisms that regulate localization of a DNA double-strand break to the nuclear periphery. Genes Dev. 2009;23:912–927. doi: 10.1101/gad.1782209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kalocsay M, Hiller NJ, Jentsch S. Chromosome-wide Rad51 spreading and SUMO-H2A.Z-dependent chromosome fixation in response to a persistent DNA double-strand break. Mol Cell. 2009;33:335–343. doi: 10.1016/j.molcel.2009.01.016. [DOI] [PubMed] [Google Scholar]
- 23.Nagai S, Dubrana K, Tsai-Pflugfelder M, Davidson MB, Roberts TM, Brown GW, et al. Functional targeting of DNA damage to a nuclear pore-associated SUMO-dependent ubiquitin ligase. Science. 2008;322:597–602. doi: 10.1126/science.1162790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dubrana K, van Attikum H, Hediger F, Gasser SM. The processing of double-strand breaks and binding of single-strand-binding proteins RPA and Rad51 modulate the formation of ATR-kinase foci in yeast. J Cell Sci. 2007;120:4209–4220. doi: 10.1242/jcs.018366. [DOI] [PubMed] [Google Scholar]
- 25.Abdallah P, Luciano P, Runge KW, Lisby M, Geli V, Gilson E, et al. A two-step model for senescence triggered by a single critically short telomere. Nat Cell Biol. 2009;11:988–993. doi: 10.1038/ncb1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Evans SK, Lundblad V. Est1 and Cdc13 as comediators of telomerase access. Science. 1999;286:117–120. doi: 10.1126/science.286.5437.117. [DOI] [PubMed] [Google Scholar]
- 27.Peterson SE, Stellwagen AE, Diede SJ, Singer MS, Haimberger ZW, Johnson CO, et al. The function of a stem-loop in telomerase RNA is linked to the DNA repair protein Ku. Nat Genet. 2001;27:64–67. doi: 10.1038/83778. [DOI] [PubMed] [Google Scholar]
- 28.Loeillet S, Palancade B, Cartron M, Thierry A, Richard GF, Dujon B, et al. Genetic network interactions among replication, repair and nuclear pore deficiencies in yeast. DNA Repair (Amst) 2005;4:459–468. doi: 10.1016/j.dnarep.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 29.Therizols P, Fairhead C, Cabal GG, Genovesio A, Olivo-Marin JC, Dujon B, et al. Telomere tethering at the nuclear periphery is essential for efficient DNA double strand break repair in subtelomeric region. J Cell Biol. 2006;172:189–199. doi: 10.1083/jcb.200505159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Palancade B, Doye V. Sumoylating and desumoylating enzymes at nuclear pores: underpinning their unexpected duties? Trends Cell Biol. 2008;18:174–183. doi: 10.1016/j.tcb.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 31.Gartenberg MR. Life on the edge: telomeres and persistent DNA breaks converge at the nuclear periphery. Genes Dev. 2009;23:1027–1031. doi: 10.1101/gad.1805309. [DOI] [PMC free article] [PubMed] [Google Scholar]