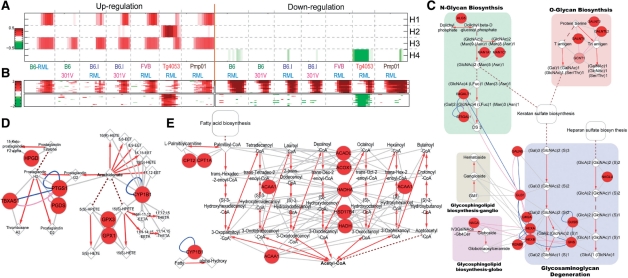
Fig. 4.
Application of PNA to gene expression data from prion-infected tissues. The results show four strain-combination-dependent activation patterns (A), as well as differential expression patterns of top 20 genes in each basis (B). Both PS (Supplementary Figure S9) and PMS (Supplementary Figure S10) for basis 1 (early PrPsc accumulation) using the significant nodes (red) and edges were reconstructed. Three pathways of the PMS (GAGs, fatty acids and arachidonates to prostaliandins; C–E), previously reported to be associated with PrPsc accumulation are shown. The round, diamond and octagon nodes indicate proteins, metabolitess and glycans, respectively. Red arrows indicate metabolic reactions, and gray edges indicate interactions between enzymes and either substrates or products. See the text for details.