Introduction
Melanoma is the most aggressive form of skin cancer. Unfortunately, despite recent improvements for some solid tumors, the prevalence and mortality of melanoma continue to increase. For patients with distant metastases, treatment with single-agent or combination chemotherapy regimens have generally resulted in very low response rates, with no significant impact on patient survival 1. Immunotherapies (i.e. interleukin-2, ipilimumab) have also yielded overall low response rates, although a small subset of patients have achieved durable responses and long-term survival 2–5. These modest achievements are further limited by noting that immunotherapies may also result in significant toxicities, including treatment-related deaths. Thus, there is a critical need for new therapeutic approaches for this aggressive disease.
The treatment of many cancers is entering into a new era based on an improved understanding of the molecular pathogenesis of these diseases. While some cancers appear to be primarily driven by viral infection, the majority are caused by genetic events that alter the expression and/or function of normal genes and proteins. These events, which include gene amplifications, deletions, and mutations, disrupt the regulatory processes that normally control the growth and survival of cells. Multiple analyses have shown that while there is a spectrum of genetic abnormalities in cancer cells, most of them affect certain signaling pathways and functions. Cancer cells are subsequently often critically dependent upon these pathways for survival, a phenomenon termed ‘oncogene addiction.’ This reliance on pathways that are hyperactivated by genetic events that occur specifically in cancer cells presents a therapeutic opportunity to block those targets in order to inhibit the growth and survival of the cancer while sparing the normal cells of the body. This approach, termed ‘targeted therapy,’ has been shown to be an effective and FDA-approved strategy for a number of cancers, including chronic myelogenous leukemia (CML), gastrointestinal stromal tumors (GIST), and renal cell carcinoma (RCC) 6. Targeted therapies have also proven effective in treating specific subpopulations of patients with other cancers, such as the use of trastuzumab (Herceptin) for HER2/neu-amplified breast cancer 7. For each of these examples, successful implementation of a targeted therapeutic approach was critically dependent upon the identification of activating genetic events, as well as the affected pathways that were present for each specific tumor type.
There is now evidence that the majority of melanomas harbor one or more mutations in critical kinase signaling pathways. Interestingly, accumulating data supports that the prevalence of these events varies greatly among the subtypes of melanoma that have been defined by clinical and pathologic characteristics 8. Most cutaneous melanomas (CM) arise from melanocytes on sun-exposed skin. Exposure to ultraviolet radiation is thought to play a major causative role in these tumors. However, the role of ultraviolet radiation is less clear for cutaneous melanomas arising from relatively sun-protected sites. Examples of such melanomas include those arising on the palms and soles; these are termed acral lentiginous melanomas (ALM). Melanomas may also arise from melanocytes in the mucosa of the head and neck, the gastrointestinal tract, and the genitourinary tract. Melanomas arising in such sites are classified as mucosal melanomas (MuM), and clearly arise in the absence of exposure to ultraviolet radiation. Consistent with the hypothesis that these melanoma subtypes are caused by different factors, comparative genomic hybridization (CGH) analysis has demonstrated that these clinically-defined groups of tumors have markedly different patterns of DNA copy number changes, including subtype-specific gene amplifications and deletions 9. CMs that arise in areas with chronic sun exposure, and that have histologic evidence of chronic sun damage (CSD), also exhibit markedly different chromosomal and gene copy number changes compared to CMs without chronic sun damage. Melanomas may also arise from melanocytes in the uveal tract of the eye (iris, ciliary body, and choroid), and are referred to as uveal melanomas (UvM). These tumors are also characterized by chromosomal changes that are distinct from CMs, ALMs, and MuMs 10, 11.
The identification of activating mutations in melanoma, combined with a growing appreciation of the different pattern of genetic changes in the anatomically defined melanoma subtypes, has become the focus of a concerted effort to translate these discoveries into personalized therapeutic approaches for this disease. In this article, we will review: (1) the known mutations, amplifications, and deletions in kinase signaling pathways that have been implicated in melanoma; (2) the prevalence of these genetic events in clinicopathologically defined melanoma subtypes; and (3) the results of clinical trials that utilize targeted therapy approaches to block aberrantly activated pathways resulting from such mutations. Importantly, we will also discuss the challenges that must be overcome in order to achieve improved outcomes with targeted therapies in melanoma in the future.
BRAF
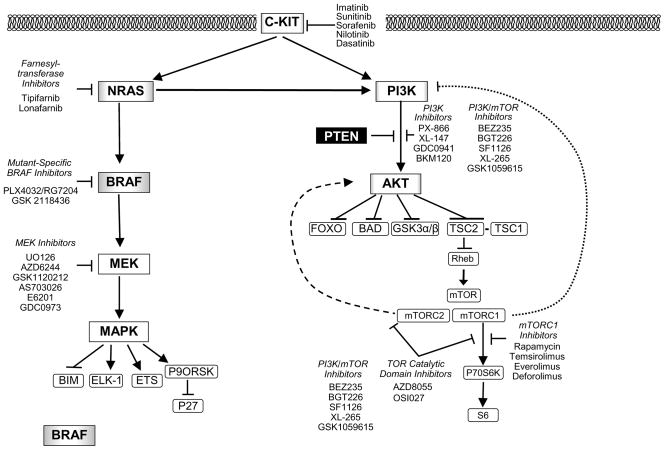
The RAS-RAF-MEK-MAPK signaling pathway is a critical regulator of cellular growth and survival [Figure 1] 12, 13. The first components of the pathway are the RAS-family GTP-ases. The RAS family members (HRAS, KRAS, NRAS) are guanine-nucleotide binding proteins that are embedded in the inner surface of the cell membrane. Normally, the RAS proteins are GDP-bound and inactive. A variety of activating signals result in the exchange of GTP for GDP, which activates RAS. RAS family members are also frequently affected by mutations in cancer that result in constitutive GTP-binding. The activated RAS members physically interact with the RAF family of serine-threonine protein kinases (ARAF, BRAF, CRAF) downstream of RAS. This interaction activates the RAF proteins, which then translocate to the cytoplasm and phosphorylate the MEK protein kinases (MEK1, MEK2). The MEK proteins are activated by this phosphorylation, and subsequently phosphorylate the P44/42 MAPK serine-threonine kinases (ERK1, ERK2). The activated MAPKs phosphorylate a variety of transcription factors and cytosolic proteins to promote proliferation and survival.
Figure 1. Kinase Signaling Pathways and Targeted Therapies for Melanoma.
The diagram illustrates key proteins in the RAS-RAF-MEK-MAPK and the PI3K-AKT kinase cascades. Arrows represent activation, while bars represent inhibition. Genes that are affected by activating mutations in melanoma (BRAF, NRAS, PI3K, C-KIT, and AKT) are shaded; the degree of shading reflects the relative prevalence of these mutations in cutaneous melanomas. Genes that are affected by genetic inactivation (PTEN) are shown with white type against a black background. The feedback regulation of PI3K and AKT by mTORC1 and mTORC2, respectively, is shown by the dashed lines. Classes and examples of targeted therapies against various effectors in the pathways are shown as free text beside the pathways.
In 2002, an experimental screen for mutations in RAF family members in cancer cell lines and tumors identified point mutations in BRAF in approximately half of the melanomas that were examined in the study, as well as occasional (3–18%) colon, lung, breast, and ovarian cancer specimens 14. Since this sentinel observation, the high frequency of point mutations in BRAF in melanoma has been confirmed in multiple studies. A recent meta-analysis of over 200 published studies reported an overall mutation rate of 65% in melanoma cell lines and 42% in tumors 15. The higher frequency of mutations in cell lines likely reflects a positive selection for cells with the BRAF mutation to propagate in vitro. Analysis of anatomic subtypes demonstrated that BRAF mutations are relatively common in CMs (42.5%), but uncommon in MuMs (5.6%) and rare in UvMs (<1%) [Table 1]. Among CMs, BRAF mutations are common in superficial spreading melanomas (SSM) (53%), but are less prevalent in acral lentiginous (ALM) (18%) and lentigo maligna melanomas (LMM) (9%) 15. LMMs are associated with CSD, and often originate in the head and neck region. The low prevalence of BRAF mutations in these tumors is consistent with the different patterns of DNA copy number gains and losses observed in the CGH analysis of cutaneous melanomas with or without chronic sun damage. BRAF mutations are also detectable in up to 80% of common acquired nevi 16–18. However, BRAF mutation rates are lower in several of the less-common nevi types, including congenital, Spitz, and blue nevi 18–20.
Table 1.
Patterns of gene mutations and amplifications in melanoma.
| Melanoma Subtype | BRAF Mutation | NRAS Mutation | c-KIT Mutation | c-KIT Amplification | GNαQ Mutation |
|---|---|---|---|---|---|
| Sun-exposed Cutaneous (CM) | 43% | 26% | Non-CSD: < 2% CSD: 2–17% |
Non-CSD: 0–7% CSD: 6% |
< 1% |
| Acral (ALM) | 18% | 4% | 18% | 24% | < 1% |
| Mucosal (MuM) | 6% | 14% | 24% | 26% | < 1% |
| Uveal (UvM) | < 1% | < 1% | < 1% | < 1% | 45% |
The table shows the prevalence of the listed genetic events in melanoma clinical specimens. CSD, chronic-sun damaged
Approximately 50 different point mutations in BRAF have been identified in cancer 21. A single substitution, the BRAF V600E mutation, comprises ~85% of the BRAF mutations detected in melanoma 15. The V600E mutation increases the in vitro kinase activity of the BRAF protein more than 400-fold 21. Most of the other reported somatic BRAF mutations, particularly other changes involving the V600 residue, also increase BRAF’s catalytic activity (5-fold to 700-fold). Interestingly, a few of the BRAF mutations that have been detected in cancer cells (G466E, G466V, G596R, D594V) decrease the catalytic activity of the BRAF protein 21, 22. As BRAF V600E is the predominant mutation identified in tumors, including melanoma, most functional studies have examined the function of the protein encoded by this change. Expression of the BRAF V600E protein results in constitutive phosphorylation and activation of MEK and MAPK 14, 21. Inhibition of BRAF V600E with small interfering RNA (siRNA, shRNA) inhibits MAPK activation, growth, and survival of human melanoma cell lines with this mutation 23, 24. These data support that melanomas with the BRAF V600E mutation depend on it for survival, and thus implicated it as a therapeutic target.
Sorafenib was the first BRAF inhibitor to be used in clinical trials in melanoma. Sorafenib is a small molecule inhibitor of multiple tyrosine kinases, including BRAF, CRAF, c-KIT, vascular endothelial growth factor receptor (VEGFR), and platelet-derived growth factor receptor (PDGFR) 25. In preclinical studies, sorafenib slowed the growth of melanoma xenografts in nude mice, but it did not cause tumor regression 26. Among 34 evaluable patients with metastatic melanoma in a phase II trial, only 1 partial response was observed 27. Subsequently, more promising results were reported in a phase I/II clinical trial of sorafenib combined with paclitaxel and carboplatin28. While the trial enrolled patients with multiple tumor types, all of the clinical responses were achieved in melanoma patients; among these patients, the response rate was 42% and the median progression-free survival was ~10 months. Although these results were encouraging when compared to previous trials in melanoma with paclitaxel and carboplatin alone (response rates of <10–20%) 29, 30, a subsequent randomized phase III trial showed that sorafenib did not improve the response rate or progression-free survival compared with the doublet of paclitaxel and carboplatin alone 31. These results raised the possibility that mutant BRAF was not a good therapeutic target in melanoma. The observation that activating BRAF mutations are present in up to 80% of benign nevi - indolent lesions with almost no malignant potential - is certainly consistent with the hypothesis that this genetic alteration alone cannot fully explain the aggressive biology of melanoma 16. Similarly, introduction of the BRAF V600E mutation alone was not sufficient to transform melanocytes into invasive lesions in multiple models, but required complementation by other genetic events to transform cells 32–34. An alternative explanation for the failure of sorafenib is that mutant BRAF is actually a good therapeutic target, but that the drug did not inhibit BRAF effectively. Interestingly, sorafenib has demonstrated marked clinical efficacy in renal cell carcinoma, a tumor characterized by dependence upon signaling by VEGFR, but not by BRAF 35, 36. Of note, in the phase II trial of paclitaxel, carboplatin, and sorafenib, there was no association noted between the presence of BRAF mutations and clinical responses, but there was a positive association with expression of the VEGFR2 protein and clinical response 28, 37.
The potential of the BRAF V600E protein as a therapeutic target for melanoma has now been validated by clinical trials with second-generation BRAF inhibitors. PLX4032 (also known as RG7204) is a more potent BRAF inhibitor than sorafenib, and it is selective for the V600E mutant form of the protein. PLX4032 inhibits the catalytic activity of the BRAF V600E protein at an IC50 of 13 nM, which is more than 10-fold lower than the dose that inhibits the wild-type protein 38. In contrast to sorafenib, an inhibitor of multiple protein kinases at therapeutic drug levels, PLX4032 has minimal activity against most other protein kinases, with an IC50 >1,000 nM for many related proteins. Preclinical studies demonstrated that PLX4032 inhibited the growth of melanoma cells with a BRAF mutation at 10x-100x lower concentrations than melanoma cell lines without a mutant BRAF 39. PLX4032 also caused the regression of BRAF-mutant melanoma xenografts in mouse models 39.
Recently, the results of a Phase I clinical trial with PLX4032 have been reported 40, 41. In the initial phase of the trial, the drug was well tolerated but no clinical responses were observed. Importantly, however, the serum levels achieved were below those that correlated with anti-tumor activity in vitro. The drug was then reformulated from a crystalline compound to a microprecipitated bulk powder. Subsequently, linear dose-dependent increases in serum levels were observed, as were clinical responses. In the Phase I trial dose expansion cohort of 32 BRAF V600E mutant melanoma patients treated with 960 mg twice daily (the dose selected for further testing), two complete responses and 24 partial responses were observed, for an overall response rate of 81% 41. The major toxicity was the development of cutaneous squamous cell carcinomas, mostly keratocanthomas, which developed in 31% of patients. These lesions were treated by surgical resection and did not result in any patient coming off study. No squamous cell carcinomas were observed at non-cutaneous sites. While this high overall response rate is unprecedented for a single agent in melanoma, several patients developed secondary resistance after their initial response noted by recurrence and/or progression of their tumors. The median duration of response to PLX4032 in the initial trial report was ~7 months 41.
The efficacy of targeting mutant BRAF is also supported by early results from the Phase I trial of GSK2118436, another potent, mutant-specific BRAF inhibitor 42. While the maximum tolerated dose has yet to be reached, patients with BRAF-mutant melanoma treated with 150 – 200 mg twice daily had a 63% response rate, and 39% of patients treated with lower doses had also responded. The duration of these responses is currently unknown. Similar to the PLX4032 trial described above, cutaneous squamous cell carcinomas also represented the major toxicity observed in the GSK2118436 trial.
The reported activities of PLX4032 and GSK2118436 in melanoma patients with BRAF V600 mutations suggests that a new standard of care will likely soon exist for these patients. However, it is also apparent that these agents should not be used in unselected melanoma patients. In the Phase I trial of PLX4032, 5 patients were included who did not have a BRAF mutation. None of these patients responded; in fact, 4 of the patients had tumor progression in the first 2 months of treatment 41. The lack of clinical response, and possible rapid progression, in patients not harboring a BRAF mutation is consistent with observations in preclinical models by four different groups suggesting potential promotion of tumor growth when mutant-selective BRAF inhibitors are used to treat BRAF wild-type melanomas 22, 43–45. In these experiments, inhibition of BRAF in melanoma cell lines expressing wild-type BRAF resulted in the hyperactivation of the MAPK pathway. While the results varied somewhat between the groups, the BRAF inhibitors promoted the formation of heterodimers of BRAF and CRAF that potently activate MEK in cells without a BRAF mutation. The cells were then dependent upon CRAF for MAPK pathway activation and survival. Interestingly, low catalytic activity BRAF mutants seem to activate MAPK similarly, and preclinical evidence suggests that melanoma cells with these mutations are sensitive to CRAF inhibition, including treatment with sorafenib 22, 46. Thus, while sorafenib failed to demonstrate activity in unselected patients, it is possible that it may be effective for certain genetic subtypes of melanoma.
NRAS
Mutations in the members of the RAS family of GTP-ases are one of the most frequent events in cancer 47. While mutations of HRAS and KRAS are common in many cancer types, they are very rare in melanoma. However, NRAS mutations have been reported in 14% of human melanoma cell lines and 15–25% of melanoma clinical specimens 15, 48–50. The mutations affecting NRAS are highly conserved; mutations affecting positions 12 and 61 constitute approximately 90% of the mutations reported in melanoma 15. The prevalence of NRAS mutations varies across the different anatomically-defined melanoma subgroups, although not as dramatically as is observed for BRAF mutations [Table 1]. NRAS mutations are detected in approximately 26% of CMs, 14% of MuMs, and <1% of UvMs 15. Among CMs, 22% of superficial spreading melanomas and 28% of nodular melanomas have NRAS mutations, while significantly lower rates are observed in acral lentiginous (4%), spitzoid (10%), and lentigo maligna (0%) melanomas 49–51. NRAS mutations are also present in common acquired nevi (6–20%) at a similar rate as has been detected in melanomas, and potentially at an even higher rate in congenital nevi 17–20. Interestingly, although rare in melanoma and other types of melanocytic nevi, HRAS mutations have been reported in 12 – 29% of Spitz nevi 51, 52.
With very rare exceptions, the common activating BRAF and NRAS mutations are mutually exclusive in melanoma tumor and cell lines 48, 49, 53. This is likely due to the fact that both of these mutations potentially activate the MAPK pathway, and the presence of both would be functionally redundant. In contrast, approximately 10% of melanomas with BRAF mutations that are catalytically inactive (i.e. D594V) also have activating NRAS mutations 22. While the activated mutant forms of BRAF and NRAS both activate MEK and MAPK, the activation of these downstream elements is CRAF-dependent in melanomas with NRAS mutations, whereas it is BRAF-dependent in BRAF-mutant cells 54.
The development of direct RAS inhibitors, a priority in a number of cancer types, has proven to be quite challenging 55. One approach has involved the development of farnesyl transferase inhibitors (FTIs), as RAS proteins must be farnesylated in order to translocate to the plasma membrane and activate their related signaling pathways. However, when farnesylation is inhibited, both NRAS and KRAS undergo an alternative modification, geranylgeranylation, that allows the proteins to be recruited to the plasma membrane 56. In addition, since over 60 cellular proteins have been shown to be farnesylated, FTIs will likely have off-target effects (and potentially dose-limiting toxicities) that compromise the ability to reach levels that effectively inhibit RAS. This lack of specificity also makes it challenging to determine the role of RAS in both the activity and toxicity of FTIs.
An alternative strategy to treat NRAS-mutant tumors is to inhibit pathways that are downstream of the mutant RAS protein. Experiments in multiple tumor types have demonstrated that mutant RAS activates multiple pro-survival and proliferative pathways in addition to the RAF-MEK-MAPK cascade 57. Other RAS effectors include PI3K, RALGDS, and PKCε. Among these, PI3K has gained particular attention, as the PI3K-AKT pathway has been implicated in melanoma by other genetic events, and it is the target of aggressive drug development [Figure 1] 58, 59.
PI3K-AKT Pathway
The PI3K-AKT pathway is one of the most important signaling networks in cancer 60. PI3K is a lipid kinase that consists of a regulatory subunit (PIK3R; p85) and a catalytic subunit (PIK3C; p110). PI3K’s catalytic activity is activated by a number of different signals, including growth factor tyrosine kinase receptors and activated RAS proteins. Activation of PI3K results in the phosphorylation of the 3′-OH of phosphatidylinositols (PI) in the plasma membrane, generating PI (3,4)P2 and PI(3,4,5)P3. These 3′-phospholipids recruit proteins to the cell membrane that have a pleckstrin homology (PH) domain, such as AKT and PDK1. AKT, a serine-threonine kinase that normally exists in an inactive state in the cytoplasm, has been extensively studied as one of the key molecules that are regulated by PI3K. Upon recruitment to the plasma membrane, AKT is phosphorylated at two critical residues, Ser473 (by the mTORC2 complex) and Thr308 (by PDK1), which activates its serine-threonine kinase activity. The activated AKT molecule translocates to the cytoplasm where it phosphorylates a variety of substrates, including FOXO, GSK3α/β, BAD, TSC2, and MDM2 [Figure 1]. Through these and other substrates, activation of AKT regulates a number of processes that contribute to the malignant phenotype, including proliferation, survival, invasion, and angiogenesis 60.
The PI3K-AKT pathway is affected by mutations that activate it more than any other signaling pathway in cancer 61. The PI3K-AKT pathway was initially implicated in melanoma by the identification of activating NRAS mutations. In addition, loss of function of PTEN, a critical negative regulator of the pathway, is a frequent event in melanoma. PTEN inhibits the activation of AKT by dephosphorylating phosphatidylinositols at the 3′ position, thereby antagonizing PI3K-mediated signaling [Figure 1] 62. Loss of PTEN results in constitutive activation of AKT in multiple cancer types, including melanoma 63. Loss of PTEN, by both genetic and epigenetic mechanisms, has been reported in 10–30% of melanomas 64–66. The prevalence of PTEN loss has been defined predominantly in cutaneous melanomas, and so the relative prevalence in anatomically-defined subtypes is not known at this time. Nonetheless, loss of PTEN is frequently detected in melanoma tumors and cell lines with a concurrent BRAF mutation, but it appears to be mutually exclusive with NRAS mutations 50, 67–69. While this pattern of mutations suggests that PTEN loss and NRAS mutations may have functional redundancy, quantitative analysis of AKT activation in melanoma tumors and cell lines showed that loss of PTEN correlated with much higher levels of activated AKT 70. This finding is similar to previous studies that showed non-equivalent activation of, and functional dependence upon, different PI3K-AKT pathway effectors by PTEN loss and PIK3CA mutations 71. P IK3CA mutations are relatively common in breast and colon cancer, but have been detected in ≤ 3% of melanomas 72, 73. Activating mutations of AKT, initially identified in breast, colon, and ovarian cancers, have also been detected as rare events in melanoma (2%) 74, 75. Each melanoma with an AKT mutation also had a BRAF mutation. While activating mutations of AKT in other cancers all involved the AKT1 isoform, some of the mutations in melanoma affected the AKT3 gene. A role for AKT3 in melanoma is supported by previous studies that showed a frequent switch from AKT1 to AKT3 expression and dependence in metastatic melanomas 76, 77.
Inhibitors against multiple components of the PI3K-AKT pathway have been developed and are in various stages of clinical testing [Figure 1] 58. Initial clinical trials were performed with mTOR inhibitors, in part because the safety of these agents had been previously established by the use of Rapamycin (an mTOR inhibitor) in transplant patients. Similar to the experience in several other cancers, mTOR inhibitors have shown little activity in melanoma. In a phase II clinical trial, the Rapamycin analog (rapalog) CCI-779 produced only one short-lived partial response among 33 patients with metastatic melanoma 78. In contrast to at least some other targeted therapies in which drug levels sufficient to significantly inhibit their intended target are not attained, it does appear that mTOR inhibitors reach levels that significantly inhibit their target in vivo 79, 80. However, studies in both clinical specimens and cell lines have demonstrated that rapalogs activate AKT, thus contributing to their lack of efficacy 80, 81. The mTOR protein participates in 2 different complexes, referred to as mTORC1 and mTORC2. The mTORC1 complex, which is inhibited by rapalogs, regulates the activation of protein translational machinery by activating P70S6K. However, it also negatively regulates PI3K as part of a feedback regulatory loop of the PI3K-AKT pathway. The mTORC2 complex, which is not inhibited by the rapalogs, phosphorylates and activates AKT 82. In preclinical models, combined inhibition of the mTORC1 and mTORC2 complexes blocked the activation of P70S6K and AKT, and more effectively inhibited growth and survival of cancer cells, than inhibition of mTORC1 alone 83. Clinical testing has yet to be reported for this agent. Similarly, clinical trials of PI3K and AKT inhibitors are ongoing. However, preclinical experiments with RAS-mutant tumors, including melanomas, showed synergistic inhibition of tumor growth and survival when inhibitors against the RAS-RAF-MEK-MAPK and the PI3K-AKT pathways were combined 84, 85. The high frequency of BRAF mutations in melanomas with PTEN loss suggests that combinatorial regimens may be necessary in other melanoma genotypes as well 86.
c-KIT
While the majority of sun-exposed cutaneous melanomas harbor an activating mutation in BRAF or NRAS in the MAPK signaling pathway, such changes are relatively rare in non-cutaneous melanomas. This disparity led to investigations that attempted to identify other genetic changes that could activate the same or other kinase signaling pathways in these tumors. In the CGH analysis of melanoma subtypes, the chromosomal region 4q12 was selectively amplified in ALMs and MuMs 9. This region harbors a number of candidate genes, but detailed analysis demonstrated that the c-KIT gene was the focal target of copy number gain in this region 87. Extra copies of c-KIT were identified in 8% of MuMs and 7% of ALMs. The c-KIT gene was also amplified in 6% of CMs with evidence of CSD, whereas no amplifications were detected in CMs without CSD. In addition, sequencing of c-KIT identified missense mutations in 21% of the mucosal, 11% of the acral, and 17% of the CSD cutaneous, but 0% of the non-CSD cutaneous melanomas 87. Subsequent studies have reported similar rates of c-KIT mutations in mucosal and acral melanomas, but they have reported lower rates of mutations in CSD cutaneous tumors, as well as different rates of gene copy number gain across the subtypes [Table 1] 88–90.
The c-KIT gene encodes a membrane tyrosine kinase receptor. Mutations of c-KIT are the most common mutation detected in gastrointestinal stromal tumors (GISTs) 91. These mutations result in constitutive activation of the c-KIT tyrosine kinase, and activation of multiple pro-survival signaling pathways, including the MAPK and PI3K-AKT pathways [Figure 1]. GISTs exhibit oncogene addiction to the mutant c-KIT proteins, and c-KIT inhibitors (e.g., imatinib) have become the standard treatment for this disease 92, 93.
The mutations that affect c-KIT in melanoma occur in the same regions of the gene as are observed in GIST. The finding of activating mutations of c-KIT in melanoma was surprising, as previous studies had demonstrated that c-KIT protein expression is frequently lost in melanoma progression 94. While c-KIT is required for normal melanocyte development, enforced expression of c-KIT in melanoma cells lines resulted in decreased growth and tumorigenicity 94. Perhaps most importantly, in three phase II clinical trials of imatinib in melanoma patients, the clinical response rate was only 1.5% 90. However, these clinical trials were overwhelmingly comprised of patients with cutaneous primary melanomas, and thus unlikely included patients with c-KIT mutations or amplifications. There are now multiple case reports of metastatic melanoma patients with c-KIT mutations achieving dramatic clinical responses to various c-KIT inhibitors 95–97. Clinical trials that are restricted to metastatic melanoma patients with c-KIT mutations or amplifications are currently ongoing.
GNαQ
Mutations in BRAF, NRAS, and c-KIT are detected in <1% of uveal melanomas [Table 1]. Recently, two different groups reported point mutations in the gene encoding the stimulatory α-subunit of G-protein coupled receptors, GNαQ 98, 99. The mutations were detected in approximately 45% of uveal melanoma clinical specimens and cell lines. The mutations were highly conserved, affecting the RAS-like domain of the protein, and specifically occur at the Q209 residue that is analogous to the Q61 residue that is frequently mutated in NRAS 99. Expression of the mutant GNαQ protein in melanocytes cooperated efficiently with other genes to induce both anchorage-independent growth and tumor formation in mice. While the pathways that are activated by and critical to the function of the GNαQ protein remain to be fully elucidated, it does appear that the RAS-RAF-MEK-MAPK pathway is one of its effectors 99. This finding suggests that inhibitors of this pathway, which were previously believed to be indicated for cutaneous melanomas only, may also have a role in uveal melanoma.
Summary
More than 10 years have passed since the FDA approved a systemic therapy for the treatment of metastatic melanoma. After years of negative clinical trials, melanoma now appears to be entering a new era in which multiple new therapeutic options will be available. The identification of clinically active targeted therapy approaches has been a gradual process, built upon an improved understanding of the appropriate use of these agents. While the recent results described here have generated great enthusiasm in the melanoma community, there clearly remain many critical hurdles to overcome.
The development of effective BRAF inhibitors, associated with unprecedented clinical response rates in appropriately selected patients, is both exciting and informative. The initial negative clinical trials with sorafenib have been followed by promising studies with PLX4032 and GSK2118436. In retrospect, the failure of sorafenib was likely due to insufficient inhibition of the MAPK pathway. Indeed, PLX4032 initially failed to demonstrate clinical activity, and only achieved clinical responses when its formulation was changed, increasing drug exposure to levels that correlated with efficacy in vitro. Thus, the successful development of this highly active therapy for BRAF-mutant melanomas was dependent upon detailed evaluation beyond simply the measurement of clinical efficacy, but also studies to determine if the drug was being appropriately dosed. Going forward, the evaluation of other experimental therapies must include assessment of the drug levels and on-target effects in order to determine if failures are due to the selection of a poor target, or alternatively are due to pharmacodynamic failure of the therapy. Melanoma presents the rare opportunity to evaluate easily accessible tumor tissue in many patients due to the frequent spread of this disease to cutaneous, subcutaneous, and lymphatic sites, and thus may be an ideal clinical venue for the evaluation of new therapies against important targets.
While targeted therapies against both BRAF and c-KIT have demonstrated marked activity in patients with mutations in these genes, secondary resistance has rapidly developed in many patients 41, 96. If the mutations or pathways that cause this resistance can be identified, rational targeted therapy combinations to overcome and/or prevent relapses may be identified. Alternatively, targeted therapies may need to be combined with other therapeutic modalities to improve outcomes. For example, while targeted therapies are seemingly characterized by high response rates but relatively short duration of response, immunotherapies are characterized by infrequent responses that are often durable. It is reasonable to hope that combining these and other modalities may lead to treatments with high rates of durable responses. Indeed, evidence exists that targeting activated pathways in melanoma may enhance the immunologic response to the tumors 100. This suggests that evaluating the effects of new therapies on the interaction of tumor cells with, and their effects on, the immune response may be important to study 101.
The discovery of BRAF, NRAS, PTEN, and c-KIT alterations in melanoma has supported the development of a variety of rational therapeutic approaches. While tremendous effort is being focused on the optimization of targeted therapies against these proteins and related pathways, approximately 30% of melanoma patients have no detectable abnormality in these genes. In order to improve outcomes in these patients, it will be critical to determine if their tumors are activating similar pathways by as yet unidentified genetic alterations or if they are characterized by dependence on completely separate and heretofore underappreciated signaling cascades in this disease.
Recently, the first whole-genome sequence of a melanoma was published 102. This study identified over 33,000 changes in the melanoma genome as compared to the germline, including almost 200 non-synonymous coding region substitutions. A second high-throughput study to identify mutations in protein tyrosine kinase family members identified somatic mutations in 19 members of this family alone 103. These initial findings suggest that identifying critical mutational events will require the sequencing of many melanomas to identify recurrent events that are most likely to be functional, which will then need to be investigated further. However, as the identification of c-KIT mutations has demonstrated, such analyses will need to incorporate the recognition of the possible molecular diversity of melanomas arising from different anatomic sites. As the recent experiences with targeted therapies have demonstrated, such investment in the understanding of the molecular biology of this disease may rapidly translate into improved outcomes in patients with this highly aggressive disease.
Acknowledgments
This work was supported in part by The University of Texas MD Anderson Cancer Center Melanoma SPORE (P50 CA93459); the American Society of Clinical Oncology Career Development Award (MAD); .MD Anderson Physician-Scientist Award (MAD); Melanoma Research Alliance Young Investigator Award (MAD); Grossman Family Foundation (JEG).
Footnotes
Disclosures: M. Davies receives research support from GlaxoSmithKline, Merck, and AstraZeneca
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Tsao H, Atkins MB, Sober AJ. Management of cutaneous melanoma. N Engl J Med. 2004;351(10):998–1012. doi: 10.1056/NEJMra041245. [DOI] [PubMed] [Google Scholar]
- 2.Atkins MB, Kunkel L, Sznol M, et al. High-dose recombinant interleukin-2 therapy in patients with metastatic melanoma: long-term survival update. Cancer J Sci Am. 2000;6 (Suppl 1):S11–14. [PubMed] [Google Scholar]
- 3.Phan GQ, Attia P, Steinberg SM, et al. Factors associated with response to high-dose interleukin-2 in patients with metastatic melanoma. J Clin Oncol. 2001;19(15):3477–3482. doi: 10.1200/JCO.2001.19.15.3477. [DOI] [PubMed] [Google Scholar]
- 4.Hodi FS, O’Day SJ, McDermott DF, et al. Improved Survival with Ipilimumab in Patients with Metastatic Melanoma. N Engl J Med. 2010 doi: 10.1056/NEJMoa1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Atkins MB, Lotze MT, Dutcher JP, et al. High-Dose Recombinant Interleukin 2 Therapy for Patients With Metastatic Melanoma: Analysis of 270 Patients Treated Between 1985 and 1993. J Clin Oncol. 1999;17(7):2105. doi: 10.1200/JCO.1999.17.7.2105. [DOI] [PubMed] [Google Scholar]
- 6.Davies M, Hennessy B, Mills GB. Point mutations of protein kinases and individualised cancer therapy. Expert opinion on pharmacotherapy. 2006;7(16):2243–2261. doi: 10.1517/14656566.7.16.2243. [DOI] [PubMed] [Google Scholar]
- 7.Mass RD, Press MF, Anderson S, et al. Evaluation of clinical outcomes according to HER2 detection by fluorescence in situ hybridization in women with metastatic breast cancer treated with trastuzumab. Clinical breast cancer. 2005;6(3):240–246. doi: 10.3816/CBC.2005.n.026. [DOI] [PubMed] [Google Scholar]
- 8.Miller AJ, Mihm MC., Jr Melanoma. N Engl J Med. 2006;355(1):51–65. doi: 10.1056/NEJMra052166. [DOI] [PubMed] [Google Scholar]
- 9.Curtin JA, Fridlyand J, Kageshita T, et al. Distinct sets of genetic alterations in melanoma. N Engl J Med. 2005;353(20):2135–2147. doi: 10.1056/NEJMoa050092. [DOI] [PubMed] [Google Scholar]
- 10.Harbour JW. Molecular Prognostic Testing and Individualized Patient Care in Uveal Melanoma. American Journal of Ophthalmology. 2009;148(6):823–829. e821. doi: 10.1016/j.ajo.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Landreville S, Agapova OA, Harbour JW. Emerging insights into the molecular pathogenesis of uveal melanoma. Future Oncol. 2008;4(5):629–636. doi: 10.2217/14796694.4.5.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Colicelli J. Human RAS superfamily proteins and related GTPases. Sci STKE. 2004;2004(250):RE13. doi: 10.1126/stke.2502004re13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rubinfeld H, Seger R. The ERK cascade: a prototype of MAPK signaling. Mol Biotechnol. 2005;31(2):151–174. doi: 10.1385/MB:31:2:151. [DOI] [PubMed] [Google Scholar]
- 14.Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417(6892):949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 15.Hocker T, Tsao H. Ultraviolet radiation and melanoma: a systematic review and analysis of reported sequence variants. Hum Mutat. 2007;28(6):578–588. doi: 10.1002/humu.20481. [DOI] [PubMed] [Google Scholar]
- 16.Pollock PM, Harper UL, Hansen KS, et al. High frequency of BRAF mutations in nevi. Nat Genet. 2003;33(1):19–20. doi: 10.1038/ng1054. [DOI] [PubMed] [Google Scholar]
- 17.Poynter JNa, Elder JTbci, Fullen DRd, et al. BRAF and NRAS mutations in melanoma and melanocytic nevi. Melanoma Research. 2006;16(4):267–273. doi: 10.1097/01.cmr.0000222600.73179.f3. [DOI] [PubMed] [Google Scholar]
- 18.Indsto JO, Kumar S, Wang L, et al. Low prevalence of RAS-RAF-activating mutations in Spitz melanocytic nevi compared with other melanocytic lesions. Journal of cutaneous pathology. 2007;34(6):448–455. doi: 10.1111/j.1600-0560.2006.00646.x. [DOI] [PubMed] [Google Scholar]
- 19.Bauer J, Curtin JA, Pinkel D, et al. Congenital melanocytic nevi frequently harbor NRAS mutations but no BRAF mutations. J Invest Dermatol. 2007;127(1):179–182. doi: 10.1038/sj.jid.5700490. [DOI] [PubMed] [Google Scholar]
- 20.Saldanha G, Purnell D, Fletcher A, et al. High BRAF mutation frequency does not characterize all melanocytic tumor types. International journal of cancer. 2004;111(5):705–710. doi: 10.1002/ijc.20325. [DOI] [PubMed] [Google Scholar]
- 21.Garnett MJ, Rana S, Paterson H, et al. Wild-type and mutant B-RAF activate C-RAF through distinct mechanisms involving heterodimerization. Molecular cell. 2005;20(6):963–969. doi: 10.1016/j.molcel.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 22.Heidorn SJ, Milagre C, Whittaker S, et al. Kinase-Dead BRAF and Oncogenic RAS Cooperate to Drive Tumor Progression through CRAF. Cell. 2010;140(2):209–221. doi: 10.1016/j.cell.2009.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hingorani SR, Jacobetz MA, Robertson GP, et al. Suppression of BRAF(V599E) in human melanoma abrogates transformation. Cancer Res. 2003;63(17):5198–5202. [PubMed] [Google Scholar]
- 24.Sumimoto H, Miyagishi M, Miyoshi H, et al. Inhibition of growth and invasive ability of melanoma by inactivation of mutated BRAF with lentivirus-mediated RNA interference. Oncogene. 2004;23(36):6031–6039. doi: 10.1038/sj.onc.1207812. [DOI] [PubMed] [Google Scholar]
- 25.Strumberg D. Preclinical and clinical development of the oral multikinase inhibitor sorafenib in cancer treatment. Drugs Today (Barc) 2005;41(12):773–784. doi: 10.1358/dot.2005.41.12.937959. [DOI] [PubMed] [Google Scholar]
- 26.Karasarides M, Chiloeches A, Hayward R, et al. B-RAF is a therapeutic target in melanoma. Oncogene. 2004;23(37):6292–6298. doi: 10.1038/sj.onc.1207785. [DOI] [PubMed] [Google Scholar]
- 27.Eisen T, Ahmad T, Flaherty KT, et al. Sorafenib in advanced melanoma: a Phase II randomised discontinuation trial analysis. Br J Cancer. 2006;95(5):581–586. doi: 10.1038/sj.bjc.6603291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Flaherty KT, Schiller J, Schuchter LM, et al. A Phase I Trial of the Oral, Multikinase Inhibitor Sorafenib in Combination with Carboplatin and Paclitaxel. Clinical Cancer Research. 2008;14(15):4836–4842. doi: 10.1158/1078-0432.CCR-07-4123. [DOI] [PubMed] [Google Scholar]
- 29.Hodi FS, Soiffer RJ, Clark J, et al. Phase II study of paclitaxel and carboplatin for malignant melanoma. Am J Clin Oncol. 2002;25(3):283–286. doi: 10.1097/00000421-200206000-00016. [DOI] [PubMed] [Google Scholar]
- 30.Zimpfer-Rechner C, Hofmann U, Figl R, et al. Randomized phase II study of weekly paclitaxel versus paclitaxel and carboplatin as second-line therapy in disseminated melanoma: a multicentre trial of the Dermatologic Co-operative Oncology Group (DeCOG) Melanoma Res. 2003;13(5):531–536. doi: 10.1097/00008390-200310000-00012. [DOI] [PubMed] [Google Scholar]
- 31.Hauschild A, Agarwala SS, Trefzer U, et al. Results of a Phase III, Randomized, Placebo-Controlled Study of Sorafenib in Combination With Carboplatin and Paclitaxel As Second-Line Treatment in Patients With Unresectable Stage III or Stage IV Melanoma. J Clin Oncol. 2009;27(17):2823–2830. doi: 10.1200/JCO.2007.15.7636. [DOI] [PubMed] [Google Scholar]
- 32.Dankort D, Curley DP, Cartlidge RA, et al. Braf(V600E) cooperates with Pten loss to induce metastatic melanoma. Nat Genet. 2009;41(5):544–552. doi: 10.1038/ng.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Michaloglou C, Vredeveld LC, Soengas MS, et al. BRAFE600-associated senescence-like cell cycle arrest of human naevi. Nature. 2005;436(7051):720–724. doi: 10.1038/nature03890. [DOI] [PubMed] [Google Scholar]
- 34.Patton EE, Widlund HR, Kutok JL, et al. BRAF mutations are sufficient to promote nevi formation and cooperate with p53 in the genesis of melanoma. Curr Biol. 2005;15(3):249–254. doi: 10.1016/j.cub.2005.01.031. [DOI] [PubMed] [Google Scholar]
- 35.Escudier B, Szczylik C, Eisen T, et al. Randomized Phase III trial of the Raf kinase and VEGFR inhibitor sorafenib (BAY 43–9006) in patients with advanced renal cell carcinoma (RCC). Paper presented at: 2005 ASCO Annual Meeting; 2005; Orlando, FL. [Google Scholar]
- 36.Ratain MJ, Eisen T, Stadler WM, et al. Phase II placebo-controlled randomized discontinuation trial of sorafenib in patients with metastatic renal cell carcinoma. J Clin Oncol. 2006;24(16):2505–2512. doi: 10.1200/JCO.2005.03.6723. [DOI] [PubMed] [Google Scholar]
- 37.Jilaveanu L, Zito C, Lee SJ, et al. Expression of sorafenib targets in melanoma patients treated with carboplatin, paclitaxel and sorafenib. Clin Cancer Res. 2009;15(3):1076–1085. doi: 10.1158/1078-0432.CCR-08-2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsai J, Lee JT, Wang W, et al. Discovery of a selective inhibitor of oncogenic B-Raf kinase with potent antimelanoma activity. Proc Natl Acad Sci U S A. 2008;105(8):3041–3046. doi: 10.1073/pnas.0711741105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang H, Higgins B, Kolinsky K, et al. RG7204 (PLX4032), a Selective BRAFV600E Inhibitor, Displays Potent Antitumor Activity in Preclinical Melanoma Models. Cancer Research. 2010;70(13):5518–5527. doi: 10.1158/0008-5472.CAN-10-0646. [DOI] [PubMed] [Google Scholar]
- 40.Flaherty KT, Puzanov J, Sosman J, et al. Phase I study of PLX4032: proof of concept for V600E BRAF mutation as a therapeutic target in human cancer. J Clin Oncol. 2009;27(15s):9000. [Google Scholar]
- 41.Flaherty KT, Puzanov I, Kim KB, et al. Inhibition of Mutated, Activated BRAF in Metastatic Melanoma. New England Journal of Medicine. 2010;363(9):809–819. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kefford RF, Arkenau H, Brown MP, et al. Phase I/II study of GSK2118436, a selective inhibitor of oncogenic mutant BRAF kinase, in patients with metastatic melanoma and other solid tumors. J Clin Oncol. 2010;28(15s):8503. [Google Scholar]
- 43.Halaban R, Zhang W, Bacchiocchi A, et al. PLX4032, a Selective BRAF V600E Kinase Inhibitor, Activates the ERK Pathway and Enhances Cell Migration and Proliferation of BRAF WT Melanoma Cells. Pigment cell & melanoma research. 2010;9999(999A) doi: 10.1111/j.1755-148X.2010.00685.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Poulikakos PI, Zhang C, Bollag G, et al. RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wild-type BRAF. Nature. 2010;464(7287):427–430. doi: 10.1038/nature08902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hatzivassiliou G, Song K, Yen I, et al. RAF inhibitors prime wild-type RAF to activate the MAPK pathway and enhance growth. Nature. 2010 doi: 10.1038/nature08833. [DOI] [PubMed] [Google Scholar]
- 46.Smalley KS, Xiao M, Villanueva J, et al. CRAF inhibition induces apoptosis in melanoma cells with non-V600E BRAF mutations. Oncogene. 2009;28(1):85–94. doi: 10.1038/onc.2008.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Giehl K. Oncogenic Ras in tumour progression and metastasis. Biol Chem. 2005;386(3):193–205. doi: 10.1515/BC.2005.025. [DOI] [PubMed] [Google Scholar]
- 48.Edlundh-Rose Ea, Egyhazi Sb, Omholt Kb, et al. NRAS and BRAF mutations in melanoma tumours in relation to clinical characteristics: a study based on mutation screening by pyrosequencing. Melanoma Research. 2006;16(6):471–478. doi: 10.1097/01.cmr.0000232300.22032.86. [DOI] [PubMed] [Google Scholar]
- 49.Omholt K, Platz A, Kanter L, et al. NRAS and BRAF Mutations Arise Early during Melanoma Pathogenesis and Are Preserved throughout Tumor Progression. Clinical Cancer Research. 2003;9(17):6483–6488. [PubMed] [Google Scholar]
- 50.Tsao H, Goel V, Wu H, et al. Genetic Interaction Between NRAS and BRAF Mutations and PTEN//MMAC1 Inactivation in Melanoma. J Investig Dermatol. 2004;122(2):337–341. doi: 10.1046/j.0022-202X.2004.22243.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bastian BC, LeBoit PE, Pinkel D. Mutations and Copy Number Increase of HRAS in Spitz Nevi with Distinctive Histopathological Features. Am J Pathol. 2000;157(3):967–972. doi: 10.1016/S0002-9440(10)64609-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van Dijk MCRFMD, Bernsen MRP, Ruiter DJP. Analysis of Mutations in B-RAF, N-RAS, and H-RAS Genes in the Differential Diagnosis of Spitz Nevus and Spitzoid Melanoma. American Journal of Surgical Pathology. 2005;29(9):1145–1151. doi: 10.1097/01.pas.0000157749.18591.9e. [DOI] [PubMed] [Google Scholar]
- 53.Greene VR, Johnson MM, Grimm EA, et al. Frequencies of NRAS and BRAF mutations increase from the radial to the vertical growth phase in cutaneous melanoma. J Invest Dermatol. 2009;129(6):1483–1488. doi: 10.1038/jid.2008.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dumaz N, Hayward R, Martin J, et al. In Melanoma, RAS Mutations Are Accompanied by Switching Signaling from BRAF to CRAF and Disrupted Cyclic AMP Signaling. Cancer Res. 2006;66(19):9483–9491. doi: 10.1158/0008-5472.CAN-05-4227. [DOI] [PubMed] [Google Scholar]
- 55.Konstantinopoulos PA, Karamouzis MV, Papavassiliou AG. Post-translational modifications and regulation of the RAS superfamily of GTPases as anticancer targets. Nature reviews. 2007;6(7):541–555. doi: 10.1038/nrd2221. [DOI] [PubMed] [Google Scholar]
- 56.Zhang FL, Kirschmeier P, Carr D, et al. Characterization of Ha-ras, N-ras, Ki-Ras4A, and Ki-Ras4B as in vitro substrates for farnesyl protein transferase and geranylgeranyl protein transferase type I. J Biol Chem. 1997;272(15):10232–10239. doi: 10.1074/jbc.272.15.10232. [DOI] [PubMed] [Google Scholar]
- 57.Downward J. PI 3-kinase, Akt and cell survival. Seminars in cell & developmental biology. 2004;15(2):177–182. doi: 10.1016/j.semcdb.2004.01.002. [DOI] [PubMed] [Google Scholar]
- 58.Courtney KD, Corcoran RB, Engelman JA. The PI3K Pathway As Drug Target in Human Cancer. J Clin Oncol. 2010 doi: 10.1200/JCO.2009.25.3641. JCO.2009.2025.3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hennessy BT, Smith DL, Ram PT, et al. Exploiting the PI3K/AKT pathway for cancer drug discovery. Nature reviews. 2005;4(12):988–1004. doi: 10.1038/nrd1902. [DOI] [PubMed] [Google Scholar]
- 60.Vivanco I, Sawyers CL. The phosphatidylinositol 3-Kinase AKT pathway in human cancer. Nat Rev Cancer. 2002;2(7):489–501. doi: 10.1038/nrc839. [DOI] [PubMed] [Google Scholar]
- 61.Yuan TL, Cantley LC. PI3K pathway alterations in cancer: variations on a theme. Oncogene. 2008;27(41):5497–5510. doi: 10.1038/onc.2008.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Maehama T, Dixon JE. The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. J Biol Chem. 1998;273(22):13375–13378. doi: 10.1074/jbc.273.22.13375. [DOI] [PubMed] [Google Scholar]
- 63.Stahl JM, Cheung M, Sharma A, et al. Loss of PTEN promotes tumor development in malignant melanoma. Cancer Res. 2003;63(11):2881–2890. [PubMed] [Google Scholar]
- 64.Mirmohammadsadegh A, Marini A, Nambiar S, et al. Epigenetic silencing of the PTEN gene in melanoma. Cancer Res. 2006;66(13):6546–6552. doi: 10.1158/0008-5472.CAN-06-0384. [DOI] [PubMed] [Google Scholar]
- 65.Wu H, Goel V, Haluska FG. PTEN signaling pathways in melanoma. Oncogene. 2003;22(20):3113–3122. doi: 10.1038/sj.onc.1206451. [DOI] [PubMed] [Google Scholar]
- 66.Zhou X-P, Gimm O, Hampel H, et al. Epigenetic PTEN Silencing in Malignant Melanomas without PTEN Mutation. Am J Pathol. 2000;157(4):1123–1128. doi: 10.1016/S0002-9440(10)64627-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Goel VK, Lazar AJ, Warneke CL, et al. Examination of mutations in BRAF, NRAS, and PTEN in primary cutaneous melanoma. J Invest Dermatol. 2006;126(1):154–160. doi: 10.1038/sj.jid.5700026. [DOI] [PubMed] [Google Scholar]
- 68.Tsao H, Zhang X, Benoit E, et al. Identification of PTEN/MMAC1 alterations in uncultured melanomas and melanoma cell lines. Oncogene. 1998;16(26):3397–3402. doi: 10.1038/sj.onc.1201881. [DOI] [PubMed] [Google Scholar]
- 69.Tsao H, Zhang X, Fowlkes K, et al. Relative Reciprocity of NRAS and PTEN/MMAC1 Alterations in Cutaneous Melanoma Cell Lines. Cancer Res. 2000;60(7):1800–1804. [PubMed] [Google Scholar]
- 70.Davies MA, Stemke-Hale K, Lin E, et al. Integrated Molecular and Clinical Analysis of AKT Activation in Metastatic Melanoma. Clin Cancer Res. 2009;15(24):7538–7546. doi: 10.1158/1078-0432.CCR-09-1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vasudevan KM, Barbie DA, Davies MA, et al. AKT-independent signaling downstream of oncogenic PIK3CA mutations in human cancer. Cancer Cell. 2009;16(1):21–32. doi: 10.1016/j.ccr.2009.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Omholt K, Krockel D, Ringborg U, et al. Mutations of PIK3CA are rare in cutaneous melanoma. Melanoma Res. 2006;16(2):197–200. doi: 10.1097/01.cmr.0000200488.77970.e3. [DOI] [PubMed] [Google Scholar]
- 73.Curtin JA, Stark MS, Pinkel D, et al. PI3-kinase subunits are infrequent somatic targets in melanoma. J Invest Dermatol. 2006;126(7):1660–1663. doi: 10.1038/sj.jid.5700311. [DOI] [PubMed] [Google Scholar]
- 74.Carpten JD, Faber AL, Horn C, et al. A transforming mutation in the pleckstrin homology domain of AKT1 in cancer. Nature. 2007;448(7152):439–444. doi: 10.1038/nature05933. [DOI] [PubMed] [Google Scholar]
- 75.Davies MA, Stemke-Hale K, Tellez C, et al. A novel AKT3 mutation in melanoma tumours and cell lines. Br J Cancer. 2008;99(8):1265–1268. doi: 10.1038/sj.bjc.6604637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Robertson GP. Functional and therapeutic significance of Akt deregulation in malignant melanoma. Cancer Metastasis Rev. 2005;24(2):273–285. doi: 10.1007/s10555-005-1577-9. [DOI] [PubMed] [Google Scholar]
- 77.Stahl JM, Sharma A, Cheung M, et al. Deregulated Akt3 activity promotes development of malignant melanoma. Cancer Res. 2004;64(19):7002–7010. doi: 10.1158/0008-5472.CAN-04-1399. [DOI] [PubMed] [Google Scholar]
- 78.Margolin K, Longmate J, Baratta T, et al. CCI-779 in metastatic melanoma: a phase II trial of the California Cancer Consortium. Cancer. 2005;104(5):1045–1048. doi: 10.1002/cncr.21265. [DOI] [PubMed] [Google Scholar]
- 79.Kim KB, Davies MA, Papadopoulos N, et al. Phase I/II study of the combination of sorafenib and temsirolimus in patients with metastatic melanoma. J Clin Oncol. 2009;27(15s):9026. doi: 10.1158/1078-0432.CCR-11-2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tabernero J, Rojo F, Calvo E, et al. Dose- and Schedule-Dependent Inhibition of the Mammalian Target of Rapamycin Pathway With Everolimus: A Phase I Tumor Pharmacodynamic Study in Patients With Advanced Solid Tumors. J Clin Oncol. 2008;26(10):1603–1610. doi: 10.1200/JCO.2007.14.5482. [DOI] [PubMed] [Google Scholar]
- 81.O’Reilly KE, Rojo F, She QB, et al. mTOR inhibition induces upstream receptor tyrosine kinase signaling and activates Akt. Cancer Res. 2006;66(3):1500–1508. doi: 10.1158/0008-5472.CAN-05-2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sarbassov DD, Ali SM, Sengupta S, et al. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Molecular cell. 2006;22(2):159–168. doi: 10.1016/j.molcel.2006.03.029. [DOI] [PubMed] [Google Scholar]
- 83.Chresta CM, Davies BR, Hickson I, et al. AZD8055 Is a Potent, Selective, and Orally Bioavailable ATP-Competitive Mammalian Target of Rapamycin Kinase Inhibitor with In vitro and In vivo Antitumor Activity. Cancer Res. 2010;70(1):288–298. doi: 10.1158/0008-5472.CAN-09-1751. [DOI] [PubMed] [Google Scholar]
- 84.Engelman JA, Chen L, Tan X, et al. Effective use of PI3K and MEK inhibitors to treat mutant Kras G12D and PIK3CA H1047R murine lung cancers. Nat Med. 2008;14(12):1351–1356. doi: 10.1038/nm.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jaiswal BS, Janakiraman V, Kljavin NM, et al. Combined targeting of BRAF and CRAF or BRAF and PI3K effector pathways is required for efficacy in NRAS mutant tumors. PLoS One. 2009;4(5):e5717. doi: 10.1371/journal.pone.0005717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tran MA, Gowda R, Sharma A, et al. Targeting V600EB-Raf and Akt3 using nanoliposomal-small interfering RNA inhibits cutaneous melanocytic lesion development. Cancer Res. 2008;68(18):7638–7649. doi: 10.1158/0008-5472.CAN-07-6614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Curtin JA, Busam K, Pinkel D, et al. Somatic activation of KIT in distinct subtypes of melanoma. J Clin Oncol. 2006;24(26):4340–4346. doi: 10.1200/JCO.2006.06.2984. [DOI] [PubMed] [Google Scholar]
- 88.Beadling C, Jacobson-Dunlop E, Hodi FS, et al. KIT gene mutations and copy number in melanoma subtypes. Clin Cancer Res. 2008;14(21):6821–6828. doi: 10.1158/1078-0432.CCR-08-0575. [DOI] [PubMed] [Google Scholar]
- 89.Handolias D, Salemi R, Murray W, et al. Mutations in KIT occur at low frequency in melanomas arising from anatomical sites associated with chronic and intermittent sun exposure. Pigment cell & melanoma research. 2010;23:210–215. doi: 10.1111/j.1755-148X.2010.00671.x. [DOI] [PubMed] [Google Scholar]
- 90.Woodman SE, Davies MA. Targeting KIT in melanoma: A paradigm of molecular medicine and targeted therapeutics. Biochemical Pharmacology. 2010;80(5):568–574. doi: 10.1016/j.bcp.2010.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hirota S, Isozaki K, Moriyama Y, et al. Gain-of-function mutations of c-kit in human gastrointestinal stromal tumors. Science (New York, NY) 1998;279(5350):577–580. doi: 10.1126/science.279.5350.577. [DOI] [PubMed] [Google Scholar]
- 92.Blanke CD, Demetri GD, von Mehren M, et al. Long-term results from a randomized phase II trial of standard- versus higher-dose imatinib mesylate for patients with unresectable or metastatic gastrointestinal stromal tumors expressing KIT. J Clin Oncol. 2008;26(4):620–625. doi: 10.1200/JCO.2007.13.4403. [DOI] [PubMed] [Google Scholar]
- 93.Demetri GD, von Mehren M, Blanke CD, et al. Efficacy and safety of imatinib mesylate in advanced gastrointestinal stromal tumors. N Engl J Med. 2002;347(7):472–480. doi: 10.1056/NEJMoa020461. [DOI] [PubMed] [Google Scholar]
- 94.Huang S, Luca M, Gutman M, et al. Enforced c-KIT expression renders highly metastatic human melanoma cells susceptible to stem cell factor-induced apoptosis and inhibits their tumorigenic and metastatic potential. Oncogene. 1996;13(11):2339–2347. [PubMed] [Google Scholar]
- 95.Hodi FS, Friedlander P, Corless CL, et al. Major response to imatinib mesylate in KIT-mutated melanoma. J Clin Oncol. 2008;26(12):2046–2051. doi: 10.1200/JCO.2007.14.0707. [DOI] [PubMed] [Google Scholar]
- 96.Woodman SE, Trent JC, Stemke-Hale K, et al. Activity of dasatinib against L576P KIT mutant melanoma: molecular, cellular, and clinical correlates. Mol Cancer Ther. 2009;8(8):2079–2085. doi: 10.1158/1535-7163.MCT-09-0459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Quintas-Cardama A, Lazar AJ, Woodman SE, et al. Complete response of stage IV anal mucosal melanoma expressing KIT Val560Asp to the multikinase inhibitor sorafenib. Nature clinical practice. 2008;5(12):737–740. doi: 10.1038/ncponc1251. [DOI] [PubMed] [Google Scholar]
- 98.Onken MD, Worley LA, Long MD, et al. Oncogenic mutations in GNAQ occur early in uveal melanoma. Invest Ophthalmol Vis Sci. 2008;49(12):5230–5234. doi: 10.1167/iovs.08-2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Van Raamsdonk CD, Bezrookove V, Green G, et al. Frequent somatic mutations of GNAQ in uveal melanoma and blue naevi. Nature. 2009;457(7229):599–602. doi: 10.1038/nature07586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Boni A, Cogdill AP, Dang P, et al. Selective BRAFV600E Inhibition Enhances T-Cell Recognition of Melanoma without Affecting Lymphocyte Function. Cancer Research. 2010;70(13):5213–5219. doi: 10.1158/0008-5472.CAN-10-0118. [DOI] [PubMed] [Google Scholar]
- 101.López-Fauqued M, Gil R, Grueso J, et al. The dual PI3K/mTOR inhibitor PI-103 promotes immunosuppression, in vivo tumor growth and increases survival of sorafenib-treated melanoma cells. International journal of cancer. 2010;126(7):1549–1561. doi: 10.1002/ijc.24926. [DOI] [PubMed] [Google Scholar]
- 102.Pleasance ED, Cheetham RK, Stephens PJ, et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2010;463(7278):191–196. doi: 10.1038/nature08658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Prickett TD, Agrawal NS, Wei X, et al. Analysis of the tyrosine kinome in melanoma reveals recurrent mutations in ERBB4. Nat Genet. 2009;41(10):1127–1132. doi: 10.1038/ng.438. [DOI] [PMC free article] [PubMed] [Google Scholar]