FIGURE 4.
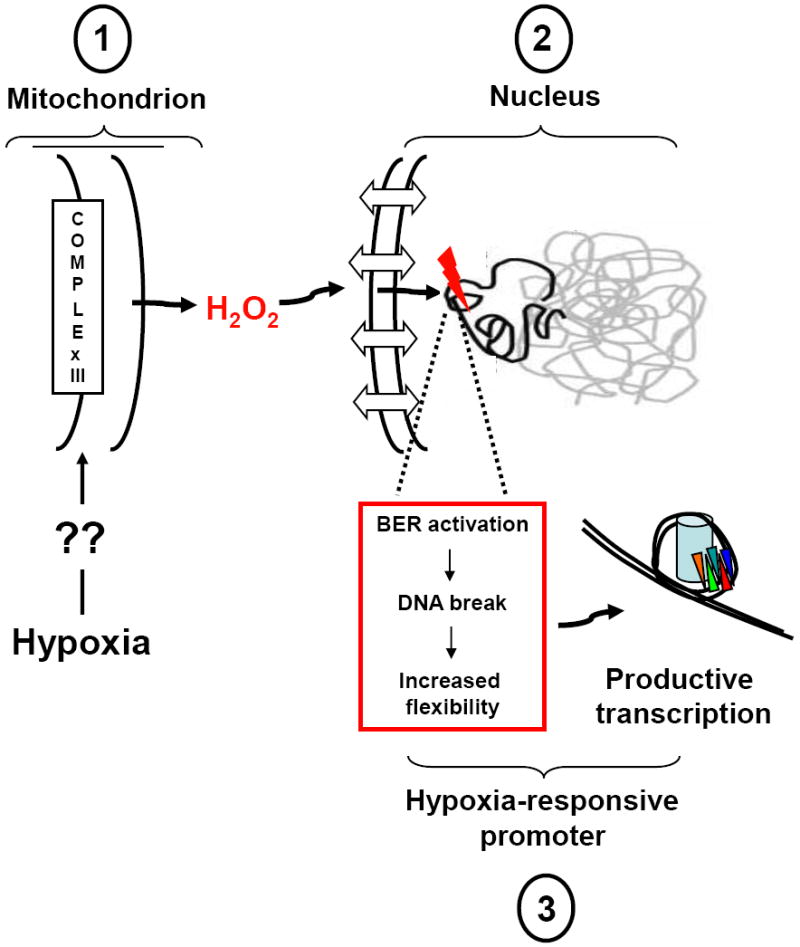
Proposed model for the role of hypoxia-generated ROS in transcriptional regulation. Key steps are: (1) By an unknown sensing and transduction mechanism, environmental hypoxia triggers mitochondrial superoxide anion production from the Uo site of complex III, thus vectoring the ROS away from the matrix-located mitochondrial genome from an oxidant stress. Superoxide anion released into the inner-membrane space is converted by resident superoxide dismutase to the comparatively long-lived signaling molecule, hydrogen peroxide, which diffuses into and accumulates in the cytosol. (2) Hydrogen peroxide generated from peri-nuclear mitochondria diffuses into the nucleus where it participates in Fenton reactions with DNA-bound iron, thus forming the highly reactive hydroxyl radical in the immediate vicinity of target DNA sequences. The reason why specific HRE sequences are targeted for oxidative base modifications is unclear, and may relate to those sequences being prepositioned in an un-protected, open conformation, to the location of DNA-bound iron in specific sequences, or to DNA charge transport which effectively tunnels ROS-mediated damage initiated elsewhere to guanine repeat sequences where the 5’ guanine is most sensitive to oxidant damage. (3) The oxidatively damaged base leads to processive recruitment of enzymes participating in the base excision pathway of DNA repair (BER). Importantly, the DNA glycosylases recognizing the oxidatively damaged base (e.g., Ogg1 in the case of oxidatively damaged purines) as well as the second enzyme in the BER pathway (Ref-1/Ape1), display strand cleaving activity that would result formation of a transient strand break in the HRE at the site of the initial oxidative damage. The strand break so produced would result in profound changes in local sequence topology and flexibility that are postulated to impact DNA-nucleosome apposition or long-range DNA conformation, thereby permitting the onset of productive transcription. Please see text for additional details and references.
