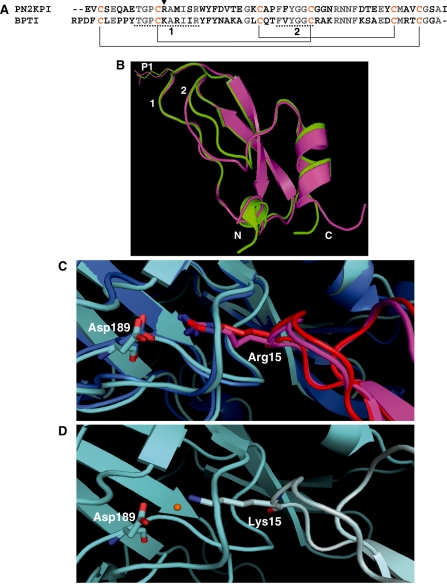
Fig. 2.
The primary and tertiary structures of the Kunitz-type inhibitors, PN2KPI and BPTI (A and B) and comparison of interactions of P1 site residues Arg15 and Lys15 (of PN2KPI and BPTI, respectively) with the Asp189 of FXIa catalytic domain and Trypsin (C and D). (A) The primary amino acid sequence of PN2KPI and BPTI. The P1 site residue is shown with an arrowhead. Underlined sequences are the enzyme interacting loops 1 and 2 of the inhibitor, which share a sequence identity of 70% and 83%, respectively. Cysteine residues that make intrachain disulfide bonds are shown in yellow and disulfide bonds are shown in solid lines. (B) Superimposed X-ray crystal structures of PN2KPI (in pink, PDB: 1ZJD) and BPTI (in green, PDB: 2FTL) shown with P1 site residues Arg15 and Lys15 side chains. Loops 1 and 2, and N and C terminals are indicated. (C) PN2KPI in complex with FXIa catalytic domain (dark blue; PDB:1ZJD) and trypsin (light blue; PDB:1TAW). Arg15 of PN2KPI (pink and red) protrudes into the S1 specificity pocket of the catalytic domain of FXIa and trypsin to establish a salt bridge with Asp189. (D) Lys15 of BPTI (grey) protrudes similarly into the S1 specificity pocket of trypsin (light blue; 2FTL) to establish a hydrogen bond with Asp189, with the intervention of a water molecule (orange).