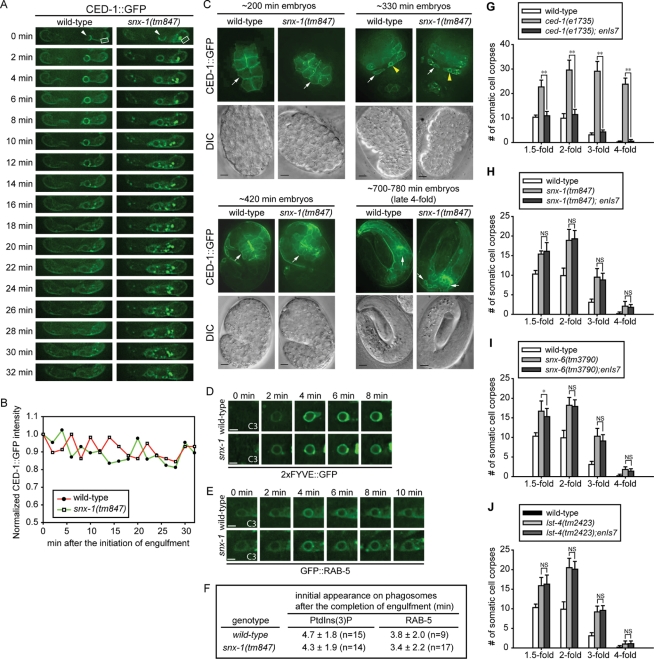
FIGURE 9:
The membrane localization of CED-1 in engulfing cells and its phagosome maturation-promoting activity are not affected by the snx mutations. (A) Time-lapse images monitoring the level of CED-1::GFP on the plasma membrane of engulfing cell ABplaapppp during and after the engulfment of cell corpse C3 (arrowhead) in wild-type and snx-1(tm847) mutant embryos. “0 min” indicates the time point when pseudopods start to extend around C3. The rectangle frames label the region from which the plasma membrane-associated GFP levels were measured and plotted in (B). (B) The fluorescence intensity of plasma membrane-associated CED-1::GFP within the framed region in (A) were measured at different time points, normalized to the GFP intensity at “0 min,” and plotted overtime. (C) CED-1::GFP is presented on the plasma membranes in snx-1(tm847) mutant embryos as in wild-type embryos at four different embryonic stages. CED-1::GFP is expressed from enIs7, an integrated transgenic array of the Pced-1ced-1::gfp reporter. White arrows indicate plasma membrane-localized CED-1::GFP. Yellow arrowheads indicate CED-1::GFP on phagosomal surfaces. Scale bar: 5 μm. (D–F) Time-lapse images monitoring the dynamic pattern of PtdIns(3)P production (D) and GFP::RAB-5 recruitment (E) on phagosomal surfaces. “0 min” represents the time point when engulfment is just completed. Scar bar: 2 μm. The quantification data of (D) and (E), obtained by monitoring multiple C1, C2, and C3 phagosomes over time, are shown in (F). n, number of phagosomes analyzed. (G–J) Overexpression of CED-1::GFP does not significantly rescue the Ced defect of snx mutant embryos. Bars represent average numbers of somatic cell corpses scored at different embryonic stages. Error bars represent standard deviation. Fifteen embryos were scored for each data point. enIs7 is an integrated transgenic array that expressed multiple copies of Pced-1 ced-1::gfp. *, **, and NS indicate p < 0.05, p < 0.01, and not significant (p > 0.05), respectively (independent Student’s t-test).