Abstract
Chronic lymphocytic leukemia (CLL), an incurable malignancy of mature B lymphocytes, involves blood, bone marrow, and secondary lymphoid organs such as the lymph nodes (LN). A role of the tissue microenvironment in the pathogenesis of CLL is hypothesized based on in vitro observations, but its contribution in vivo remains ill-defined. To elucidate the effects of tumor-host interactions in vivo, we purified tumor cells from 24 treatment-naive patients. Samples were obtained concurrently from blood, bone marrow, and/or LN and analyzed by gene expression profiling. We identified the LN as a key site in CLL pathogenesis. CLL cells in the LN showed up-regulation of gene signatures, indicating B-cell receptor (BCR) and nuclear factor-κB activation. Consistent with antigen-dependent BCR signaling and canonical nuclear factor-κB activation, we detected phosphorylation of SYK and IκBα, respectively. Expression of BCR target genes was stronger in clinically more aggressive CLL, indicating more effective BCR signaling in this subtype in vivo. Tumor proliferation, quantified by the expression of the E2F and c-MYC target genes and verified with Ki67 staining by flow cytometry, was highest in the LN and was correlated with clinical disease progression. These data identify the disruption of tumor microenvironment interactions and the inhibition of BCR signaling as promising therapeutic strategies in CLL. This study is registered at http://clinicaltrials.gov as NCT00019370.
Introduction
Chronic lymphocytic leukemia (CLL) is characterized by the progressive accumulation of mature, monoclonal B lymphocytes in the peripheral blood (PB), bone marrow (BM), and secondary lymphoid organs such as the lymph nodes (LN).1 CLL is divided into 2 main subgroups based on the presence or absence of acquired somatic mutations in the immunoglobulin heavy chain gene (IGHV) expressed by leukemic B cells. Patients whose tumor cells express an IGHV gene carrying somatic mutations (M-CLL) have a more indolent disease and longer overall survival than do patients whose tumors express an IGHV gene in the germline or “unmutated” configuration (UM-CLL). Despite important biological and clinical differences, gene expression profiling identified these 2 subtypes as part of a shared disease process with a common characteristic gene expression signature.2,3 Nevertheless, a distinct set of genes is differentially expressed between the 2 subtypes. Surprisingly, ZAP-70, a tyrosine kinase essential for T-cell receptor signaling, was the most discriminating feature between UM-CLL and M-CLL.3,4 ZAP-70 is typically expressed at higher levels in UM-CLL than in M-CLL and has become an important prognostic marker.4–7 In addition, the expression of ZAP-70 affects intracellular signaling pathways and may contribute to differences in tumor biology between the 2 CLL subtypes.8–11
Historically, CLL has been viewed as an accumulative disease of cells with a defect in apoptosis. Consistent with this view, the majority of peripheral blood CLL cells are arrested in G0/G1 and show a gene expression profile of resting cells.3 However, recent studies using deuterated water labeling indicate a more important role of tumor proliferation in the progression of CLL than was previously appreciated.12 Immunohistochemistry for the cell-cycle marker Ki67 suggests that CLL proliferation occurs in the BM and secondary lymphoid organs. The signals that govern tumor proliferation remain elusive because most in vitro systems are not able to support CLL cell proliferation.
When cultured in vitro, CLL cells rapidly undergo apoptosis, from which they can be rescued by contact with stroma cells or by the addition of soluble factors.13,14 In vitro, a wide range of different molecules can extend CLL survival, raising the specter of an opportunistic tumor that benefits from all kinds of host factors and therefore might be able to evade targeted interventions. However, in vitro systems can only extend CLL cell survival for a limited time, indicating that essential factors present in vivo are missing. Another limitation is that in vitro studies typically analyze PB-derived tumor cells because BM and LN biopsies are often not available. Thus, the contribution of the host microenvironment to the proliferation and survival of CLL cells in vivo remains ill-defined.
Chronic active BCR signaling due to point mutations in CD79b has recently been identified as a key pathogenic mechanism in aggressive B-cell lymphoma, and results in constitutive nuclear factor-κB (NF-κB) activation.15 In contrast, CLL cells have the gene expression characteristics of resting B cells, and cells from the M-CLL subtype have been described as anergic and unresponsive to BCR activation.16 While UM-CLL cells have been shown to react to immunoglobulin M (IgM) activation in vitro, evidence for BCR signaling in vivo is lacking. The BCR of many CLL cells shares characteristics with natural antibody-producing B cells that recognize microbial antigens and self-antigens, leading to the hypothesis that antigen selection plays a role in the ontogeny of CLL.17 However, where and when CLL cells respond to antigen and whether BCR activation plays a role in CLL progression have not been determined.
Gene expression profiling has made major contributions to the classification of lymphoid malignancies by dissecting biological entities based on common pathogenic pathways. In the present study, we applied gene expression profiling to investigate the effect of the microenvironment on CLL cells in vivo. To obtain a direct measure of tumor biology, we purified CLL cells simultaneously from PB, BM, and LN for gene expression profiling, which can simultaneously detect the activation of many different signaling pathways and the resulting cellular response.18 Our analysis identified signaling pathways engaged in CLL cells in the tissue microenvironment that are able to sustain CLL proliferation and survival in vivo. These data provide the framework for the development and testing of therapies that target essential pathogenic pathways in CLL.
Methods
Patients and samples
With informed consent in accordance with the Declaration of Helsinki and approval from the NIH institutional review board, PB, BM, and LN biopsies were collected from treatment-naive CLL patients (Table 1) enrolled in this National Cancer Institute Study no. 97-C-0178 (http://clinicaltrials.gov identifier: NCT00019370). Matched samples from different anatomic compartments were obtained on the same day, processed, and analyzed in parallel. Mononuclear cells were isolated by centrifugation over lymphocyte separation medium (ICN Biomedicals), followed by CD19+ selection (Miltenyi Biotec). CD19+ cells with purity > 96% were aliquoted and stored as pellets at −80°C. RNA was extracted using RNeasy kits (QIAGEN).
Table 1.
Patient characteristics
| Study number | Sex/age, y | Rai stage | IGHV gene, % germline | ZAP-70 IHC | CD38, % | FISH | BM, % cellular/pattern | LN-pc pattern | TTT, mo |
|---|---|---|---|---|---|---|---|---|---|
| C01 | F/61 | 2 | 4-59, 100 | + | 28 | +12 | 75/D | prominent | +28.5 |
| C02 | F/60 | 1 | 3-21, 96.3 | + | 88 | 11q−, 13q− | 85/D+N | typical | +79.7 |
| C03 | M/60 | 1 | 1-18, 93.9 | − | 0 | 13q− | 90/D | prominent | 106.4 |
| C04 | M/51 | 3 | 3-33, 100 | + | 53 | 11q− | 90/D | typical | +3.5 |
| C05 | M/57 | 4 | 3-15, 97.7 | − | 46 | 11q−, 13q− | na | typical | +221 |
| C06 | M/58 | 3 | 3-30, 100 | + | 60 | normal | na | prominent | +9.6 |
| C07 | F/61 | 2 | 3-21, 100 | + | 81 | normal | na | typical | +21.6 |
| C08 | M/59 | 1 | 3-09, 99.3 | + | 47 | 11q−, 13q− | 50/D+N | typical | +11.6 |
| C09 | M/39 | 3 | 1-69, 99.6 | + | 27 | +12 | 90/D | typical | +10.6 |
| C10 | M/30 | 4 | 3-09, 99.7 | + | 100 | 13q− | 80/D | prominent | +23.2 |
| C11 | F/67 | 2 | 3-30, 84.8 | − | 1 | 13q−, 17p− | 60/N | prominent | 101.9 |
| C12 | M/67 | 2 | 4-67, 94.9 | − | 3 | 13q− | 50/N | parafollicular | 29.4 |
| C13 | F/68 | 3 | 4-b, 100 | + | 97 | +12 | 80/D | diffuse | +10.5 |
| C14 | F/55 | 1 | 1-69, 100 | + | 2 | 13q− | na | typical | +35.6 |
| C16 | M/73 | 4 | 7-41, 93 | − | 1 | na | 85/D | na | +260 |
| C18 | M/56 | 2 | 3-74, 90.4 | − | 1 | normal | 40/N | na | +50.4 |
| C19 | F/73 | 1 | 3-30, 92.9 | − | 1 | 13q− | 30/N | na | 138 |
| C20 | M/73 | 1 | 4-39, 97.3 | − | 1 | 13q− | 30/N | na | 41.2 |
| C21 | F/74 | 1 | 4-04, 100 | + | 30 | 13q− | 10/D | na | 29.6 |
| C22 | M/74 | 2 | 1-18, 98.7 | + | > 30 | 13q− | 65/D+N | na | 121.2 |
| C23 | F/56 | 1 | 3-09, 98.6 | − | 68 | +12 | 75/D+N | na | 50.4 |
| C24 | M/52 | 2 | 4-61, 98.7 | + | 10 | 11q−, 17p− | na | typical | +3.9 |
| C25 | M/53 | 4 | 3-30, 99.7 | + | 72 | 13q− | 60/D+N | typical | +123 |
| C26 | F/57 | 2 | 3-09, 100 | + | 39 | +12 | 85/D | prominent | +21.7 |
IHC indicates immunohistochemistry; TTT, time to treatment (months from diagnosis to initiation of treatment); +, patients treated (treatment consisted of rituximab 375 mg/m2 on day 1 and fludarabine 25 mg/m2 on days 1-5 cycled every 4 weeks for 6 cycles; all others remained under continued observation without treatment); FISH, fluorescence in situ hybridization (only abnormalities detected in at least 10% of cells are shown); BM, bone marrow (% cellularity is given and the infiltration pattern is classified as N, nodular, or D, diffuse); LN-pc, pattern of proliferation centers in lymph node; and na, no sample available.
IGHV gene and ZAP-70 analysis
Analysis of IGHV gene status and ZAP-70 was performed as described in Wiestner et al4 ZAP-70 expression was scored on BM biopsies read by 2 independent pathologists.
Reagents and flow cytometry
Fluorescein isothiocyanate (FITC)–conjugated anti-murine IgG1 and FITC-conjugated anti-CD19 antibodies; anti-SYK antibodies; phycoerythrin (PE)–conjugated anti-murine IgG1 antibodies; PE-conjugated anti-CD69, anti-CXCR4, and p-SYK (pY348) antibodies; and PE-Cy5–conjugated CD3 antibodies were from Becton Dickinson. Anti-IκB, anti-pIκB (ser 32-36), and JunB antibodies were from Cell Signaling Technology. Anti–γ-tubulin antibodies were from Sigma-Aldrich. Anti-TATA binding protein (anti-TBP) antibodies were from Abcam. Anti-E2F1 and c-MYC antibodies were from Santa Cruz Biotechnology. For flow cytometry, 5 μL of the antibodies was added to 5 × 105 cells and incubated for 30 minutes on ice. Samples were washed with phosphate-buffered saline/1% fetal calf serum and assayed on an FC500 flow cytometer (Coulter).
Microarray hybridization, data processing, and analysis
Total RNA (1 μg) was amplified as described by the manufacturer (Affymetrix). Biotin-labeled RNA (20 μg) was fragmented to ∼ 200 bp and hybridized to U133 plus 2.0 chips for 16 hours, washed, and stained on a fluidics station. GeneChip operating software Version 1.4 was used to calculate signal intensity and present calls on the hybridized chip. The signal-intensity values obtained for probe sets in the microarrays were transformed with an adaptive variance-stabilizing, quantile-normalizing transformation (P. J. Munson, GeneLogic Workshop of Low Level Analysis of Affymetrix GeneChip Data, 2001. Software available at http://abs.cit.nih.gov/geneexpression.html). Probe sets (10 219) that had no signal in any of the 62 samples were excluded. One-way analysis of variance with blocks by patients was performed to evaluate different tissues. In the 12 patients in whom all 3 compartments had been arrayed, the patient effect on gene expression was subtracted by mean centering the expression value of each gene across the 3 compartments for each patient separately. The JMP statistical software package 7.0 (www.jmp.com, SAS Institute) and the Cluster and Tree View programs (Eisen Laboratory, Stanford University) were used. Gene Set Enrichment Analysis (GSEA, Version 2.04)19 was used to identify overrepresentation of gene sets from the online database available at the GSEA Web site (http://www.broadinstitute.org/gsea/). In addition, select gene sets from the gene expression database of the Staudt laboratory (http://lymphochip.nih.gov/signaturedb/index.html) were uploaded to GSEA for inclusion in the analysis. Enriched or overrepresented gene sets between LN and PB were identified using 1000 permutations of the phenotype labels. Primary gene expression data are deposited in Gene Expression Omnibus (GEO) under accession number GSE21029.
Western blotting
Purified CLL cells were lysed in 1% Triton buffer containing PhosSTOP and the protease inhibitor Complete, EDTA-free (Roche). Nuclear and cytosol fractions were obtained using a commercial kit (BioVision). Proteins were separated on a sodium dodecyl sulfate–acrylamide gel, transferred to polyvinylidene fluoride membranes and subjected to immunoblotting using the indicated antibodies and horseradish peroxidase–labeled secondary antibodies (Amersham). Blots were developed by chemiluminescence (Thermo Scientific), recorded on an LAS-4000 imaging system (Fujifilm), and quantified using Multi Gauge software (Version 3.1; Fujifilm).
In vitro stimulation and target gene analysis by lymphochip microarray
To identify target genes of BCR signaling, CLL cells were stimulated with goat F(ab′)2 anti–human IgM (50 μg/mL; Jackson ImmunoResearch Laboratories) at 37°C for 6 hours. Lymphochip analysis of IgM-stimulated CLL cells was conducted as described in Alizadeh et al.20
BCR activation and phosphospecific flow cytometry
CLL cells (1 × 106) were incubated with biotinylated IgM at 25 μg/mL for 30 minutes at 4°C, washed in phosphate-buffered saline, 1% fetal calf serum, 1% NMS, 0.02% NaN3 (FACS buffer), and incubated with streptavidin-APC-Cy7 (Becton Dickinson) for 10 minutes at 4°C, and then warmed to 37°C. Basal levels of phosphorylation were determined in unstimulated cells fixed at time zero. Cells were fixed for 15 minutes at room temperature in 1.4% final paraformaldehyde (Polysciences), pelleted, washed, permeabilized in 2 mL of phosphobuffer II (Becton Dickinson) for 10 minutes, and stored at 4°C. The cells were washed twice, resuspended in 100 μL of FACS buffer, and stained with PE-conjugated p-SYK (pY348), FITC-conjugated SYK, and PE-Cy5–conjugated CD3 (Becton Dickinson) for 20 minutes at room temperature. Cells were then washed twice, resuspended in 200 μL of flow buffer with 2% paraformadehyde, and analyzed on an LSRII flow cytometer (Becton Dickinson) using FACSDIVA 6.1.1 and FlowJo software (Version 8.8.6; TreeStar). A minimum of 50 000 B cells (CD3−, SYK+) were collected; BCR-activated cells were gated using APC-CY7. pSYK expression was measured as a percentage of CD3−SYK+IgM+ cells. Cutoffs for positivity were set using the fluorescence minus 1 control. To measure the expression of pSYK in cells ex vivo, H2O2 was added to a 3.3mM final concentration for 3 minutes at 37°C before fixation. Permeabilization, staining, and analysis were performed as described for BCR-stimulated cells.
Interphase fluorescence in situ hybridization
Fluorescence in situ hybridization was performed on cells cultured in duplicate overnight without mitogens and for 96 hours with B-cell mitogens, fixed in 3:1 methanol:glacial acetic acid, and hybridized with commercially available probes (Abbott Molecular, formerly Vysis) to detect deletions in 13q14.3 (D13S319, D13S25), 11q23 (MLL), 11q22.3 (ATM), and 17p13.1 (p53) and to detect trisomy 12 (CEP 12 DNA). A minimum of 200 interphase nuclei were scored for hybridization signals for each probe.
Results
The tumor biology of CLL cells in vivo is affected by the tissue microenvironment
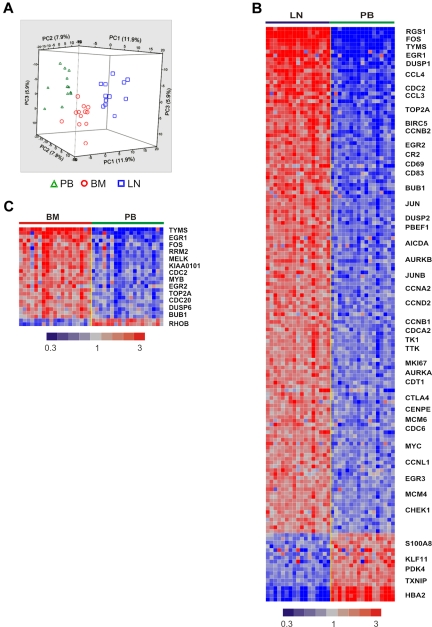
To what degree in vitro systems mirror the effect of the microenvironment on CLL cells in vivo remains unknown. To determine the influence of the physiologic tissue microenvironment on the tumor biology of CLL cells in vivo, we analyzed matched blood and tissue samples that were simultaneously obtained and immediately processed. All patients were untreated at the time of sample acquisition and represent the clinical spectrum of CLL (Table 1, supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). PB and BM were aspirated into heparinized syringes, and LN samples were obtained by surgical biopsy followed by mechanical disruption to yield single-cell suspensions. To remove variations in cellular composition of samples from different anatomic sites, we purified the tumor cells to > 96% purity using positive selection. Purified tumor cells were then analyzed on Affymetrix whole genome microarrays. Unsupervised hierarchical clustering revealed an overriding effect of sample derivation from individual patients, so that different samples aligned by patient origin (supplemental Figure 2). In 12 patients in whom cells from all 3 anatomic sites were available, gene expression was normalized to adjust for the patient effect. After normalization, unsupervised clustering of all 44 456 probe sets clearly separated the tumor cells according to the compartment of origin (Figure 1A). Thus, the gene expression complement of CLL cells is shaped by an intrinsic effect of the individual patient and an extrinsic effect determined by the tumor microenvironment.
Figure 1.
The tissue microenvironment affects the tumor biology of CLL cells in vivo. (A) Gene expression analysis of fresh CLL tumor cells derived from PB, BM, and LN in treatment-naive patients in whom all 3 sites were evaluable (n = 12). Principal component analysis of gene expression normalized for patient effect (see “Microarray hybridization, data processing, and analysis”) is shown. (B) Heat map of 151 genes differentially expressed in purified CLL cells obtained simultaneously from LN and PB in 17 patients (> 2-fold change, FDR < 20%). Patient samples are arranged in columns keeping the same order in the LN and PB groups. Gene expression is median-centered and scaled as indicated. Gene symbols highlight select genes. (C) Heat map of 26 differentially expressed genes in purified CLL cells isolated from BM aspirates compared with cells simultaneously obtained from the PB in 19 patients (> 2-fold change, FDR < 20%).
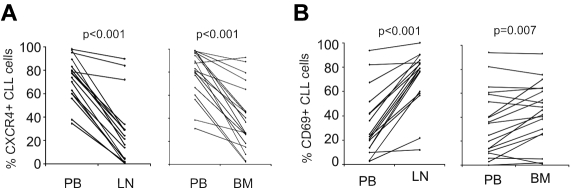
Next we compared PB-derived CLL cells against LN- and BM-derived cells using 1-way analysis of variance. CLL cells in the LN differentially expressed 151 genes, with expression levels that were at least 2-fold different between LN- and PB-derived cells (false discovery rate, FDR < 0.2, Figure 1B). Of these, 133 genes were up-regulated in LN and 18 were down-regulated compared with the PB. Using the same criteria, only 26 genes were differentially expressed between BM- and PB-derived cells, 24 being more highly expressed in BM and 2 showing reduced expression (Figure 1C). Almost all of the genes up-regulated in BM-resident cells were also up-regulated in LN-resident cells, while over 100 genes were up-regulated in the LN but not in the BM. To rule out dilution of BM aspirates with PB cells, we quantified expression of CXCR4 on CLL cells by flow cytometry. CXCR4 is internalized and down-modulated in response to binding of its cognate ligand CXCL12 (SDF-1) in the stroma environment.21 As expected, we found lower CXCR4 expression by flow cytometry on CLL cells obtained from the tissue compartments than on PB-derived cells (Figure 2A). In addition, CXCR4 cell surface expression on CLL cells in BM aspirates matched expression on cells eluted in vitro from BM core biopsies (supplemental Figure 3). These findings exclude a significant dilution of BM aspirates with PB cells. We next analyzed the expression of the activation marker CD69 on CLL cells by flow cytometry. CD69 was up-regulated on cells in the tissue microenvironment, both in BM and LN. However, consistent with the gene expression data, we also found stronger up-regulation of CD69 in LN-resident compared with BM-resident CLL cells (Figure 2B). Thus, the LN microenvironment appears to have a stronger effect on CLL cell activation than the BM microenvironment.
Figure 2.
Dynamic changes in CXCR4 and CD69 expression on CLL cells in different anatomic compartments. (A) CXCR4 was quantified by flow cytometry on CD19+-gated cells and expressed as the percentage of cells expressing CXCR4 above isotype control. Samples obtained simultaneously from the same patient are connected by a line. Mononuclear cells were obtained by density gradient centrifugation, maintained on ice, and analyzed within 24 hours. Normal B cells constituted less than 1% of CD19+ cells. Comparison was by paired t test. (B) CD69 was quantified by flow cytometry on CD19+-gated cells and expressed as the percentage of cells expressing CD69 above isotype control. Sample collection and analysis were as in panel A.
We used GSEA to investigate the biological basis of the gene expression differences in LN-resident cells. This observer-independent statistical method can dissect gene expression data into functional motifs. Because of the more pronounced difference in gene expression in LN-derived cells, we focused our analysis on this compartment. Among the genes differentially expressed in LN-resident cells, GSEA identified several significantly enriched gene sets (FDR < 0.05), indicating activation of distinct signaling pathways and tumor proliferation in the LN (Table 2).
Table 2.
Gene sets up-regulated in CLL cells in LN compared with PB
| Gene sets | Number of enriched gene sets | NES | FDR, q value |
|---|---|---|---|
| Functional gene sets | |||
| B-cell receptor signaling | 1 | 2.06 | 0.0031 |
| T-cell receptor signaling | 2 | 2.01 | 0.0043 |
| P53 pathway | 5 | 2.00 | 0.0053 |
| NF-κB pathway* | 3 | 1.98 | < 0.0001 |
| Calcineurin-NFAT signaling | 1 | 1.92 | 0.0071 |
| MYC-regulated genes | 8 | 1.89 | 0.0088 |
| Proliferation/cell cycle | 7 | 1.88 | 0.0085 |
| Pyrimidine metabolism | 2 | 1.88 | 0.0085 |
| TNF pathway | 3 | 1.88 | 0.0085 |
| CLL BCR signature† | 1 | 1.81 | 0.006 |
| Toll receptor signaling | 2 | 1.80 | 0.0151 |
| E2F1-regulated genes | 1 | 1.80 | 0.0157 |
| Motif gene sets | |||
| E2F | 19 | 2.15 | < 0.0001 |
| ATF | 1 | 1.84 | 0.006 |
| NF-Y | 2 | 1.84 | 0.006 |
| CREB | 4 | 1.85 | 0.006 |
Enriched gene sets were identified using GSEA, selected for FDR < 0.05 and NES ≥ 1.80. Where GSEA identified several gene sets with the same functional annotation, the total number of enriched gene sets is given and the NES and FDR of the most significantly enriched set is shown. Only gene sets in which at least 5 genes accounted for the enrichment score (leading edge genes) are included. Gene sets specific to nonlymphoid cell types are not shown.
NES indicates normalized enrichment score; and TNF, tumor necrosis factor.
Functional gene signatures of coordinately regulated genes derived from experimental validation: downloaded from the Staudt Lab Web site at http://lymphochip.nih.gov/signaturedb/index.html.
Functional gene signature of coordinately regulated genes derived from experimental validation: established as shown in Figure 3A.
B-cell receptor activation in LN-resident CLL cells
Many of the genes overexpressed in LN-resident cells are well recognized BCR target genes.22 This impression was confirmed by GSEA, which identified a set of BCR-related genes as the most overrepresented gene set in LN-resident CLL cells. In addition, gene sets of the NF-κB and NFAT pathways, both of which are activated by BCR signaling, were also among the top-ranked gene sets. The gene expression data thus provided a first indication for BCR activation in CLL cells in vivo.
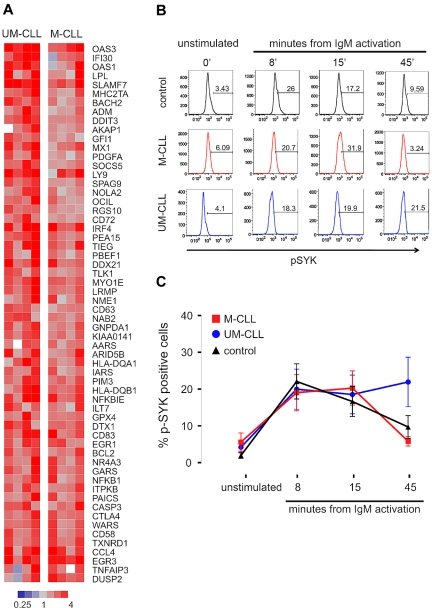
Expression of the BCR gene set did not appear to be significantly different between the 2 CLL subtypes. This was surprising, given the concept that M-CLL cells are anergized and reportedly do not respond to BCR stimulation in vitro.16 In contrast, our data suggested that in vivo, at least in the LN microenvironment, M-CLL cells do react to BCR engagement. To investigate this discrepancy, we stimulated PB CLL cells from 8 representative patients (4 UM-CLL, 4 M-CLL) using anti-IgM monoclonal antibody in vitro, and analyzed stimulation-induced gene expression changes. Across both CLL subtypes, 60 genes were up-regulated at least 2-fold (P < .01) within 6 hours of activation, and we summarized these into a “CLL-BCR signature” (Figure 3A). Almost all of the BCR target genes were more strongly up-regulated in UM-CLL than in M-CLL patients. Next we used flow cytometry to measure SYK activation in PB-derived CLL cells in response to BCR activation in vitro (Figure 3B-C). CLL cells of both subtypes and normal naive B cells showed rapid phosphorylation of SYK in response to IgM cross-linking, and at 8 and 15 minutes there was no difference between CLL cells of either subtype and normal B cells. However, at 45 minutes from BCR engagement, UM-CLL cells showed significantly higher p-SYK levels than M-CLL (P = .013). This prolonged activation of the signaling cascade in UM-CLL confirms reports by others,10 and can explain the stronger expression of BCR target genes in UM-CLL patients. Thus, PB-derived CLL cells of both subtypes are responsive to BCR engagement, activate SYK, and up-regulate BCR target genes.
Figure 3.
Activation of the B-cell receptor on CLL cells induces a characteristic gene expression signature and phosphorylation of SYK. (A) Gene expression changes in UM-CLL (n = 4) and M-CLL (n = 4) samples 6 hours after in vitro IgM cross-linking. Sixty-one genes showing a > 2-fold change between unstimulated and stimulated cells (P < .001) were identified and constitute a “CLL-BCR gene signature.” Gene expression changes in response to BCR engagement is depicted in a heat map according to the scale shown. Genes are sorted top to bottom for the ratio between the average up-regulation in UM-CLL compared with M-CLL cells. (B) Phosphorylation of SYK (p-SYK) after in vitro IgM cross-linking was assessed by flow cytometry in cells gated on CD3−SYK+IgM+ at the indicated time points. (C) Mean and SD of the percentage of pSYK-positive cells in CLL subtypes (UM-CLL n = 14, M-CLL n = 13) and normal B cells (control n = 11) are shown. Differences between groups was assessed by Student t test (P = .013 for UM-CLL vs M-CLL at 45 minutes; all other comparisons were not significant).
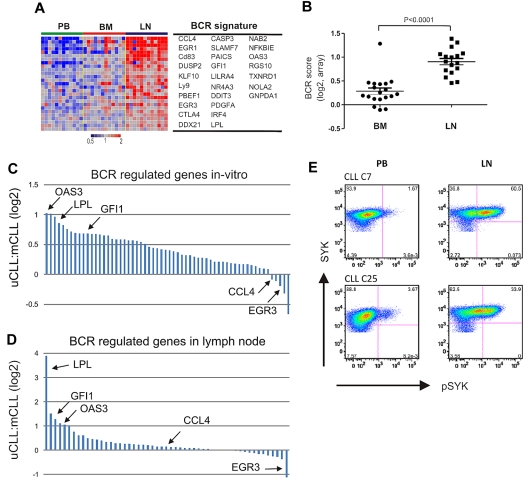
We used this functionally defined CLL-BCR signature to interrogate the gene expression of CLL cells in the microenvironment. Using GSEA, we found the CLL-BCR signature to be highly overrepresented among genes up-regulated in CLL cells in the LN (Table 2). The expression of BCR signature genes was high in the LN, low in the PB, and except for 1 patient, low in the BM (Figure 4A). To quantify the strength of BCR induction in the different anatomic sites, we computed a “BCR score” by averaging the expression level of the most highly enriched CLL-BCR signature genes. For each patient in the study, the score of tissue-derived samples was normalized to the respective PB sample (Figure 4B). All LN-derived cells showed an increase in the BCR score compared with those from the PB (n = 17, median 2.01-fold). In addition, 16 of 19 BM-derived samples had higher BCR scores than PB cells, but the increase was modest (median 1.17-fold). Thus, BCR target genes are up-regulated in CLL cells in vivo. In the vast majority of cases, this induction is stronger in the LN than in the BM.
Figure 4.
BCR activation in LN-resident CLL cells. (A) Genes in the CLL-BCR signature that were most significantly enriched in the LN as identified by GSEA (leading-edge genes) are depicted in a heat map for the 12 patients having contributed cells from all 3 compartments. (B) The BCR score was computed as the average of the mRNA expression level of the leading edge genes for each sample. Shown is the ratio of the BCR score in BM (n = 19) or LN (n = 17) relative to the score of the matched PB sample. Comparison between BM and LN was by Student t test. The sample with exceptionally high score in BM was obtained from CLL_C10. (C, D) Relative gene expression changes of BCR-regulated genes between UM-CLL and M-CLL. Each gene is represented by a bar, select genes are highlighted. (C) Ratio of relative gene expression change after IgM cross-linking between UM-CLL (n = 4) and M-CLL (n = 4). Bars represent 61 genes, as described in Figure 3A. The fold up-regulation of each gene was averaged for each subtype and the ratio between UM-CLL and M-CLL is shown on a log2 scale. (D) Ratio of relative gene expression change in UM-CLL (n = 12) and M-CLL (n = 5) between the LN- and matched PB-derived cells. Bars represent all 61 genes as in panel C. The fold up-regulation of each gene in the LN-derived cells compared with the matched PB sample was averaged within each subtype, and the ratio between UM-CLL and M-CLL is shown on a log2 scale. (E) Phosphorylation of SYK was assessed by flow cytometry in CD3− cells from the indicated anatomical site. Shown are 2 representative patients of a total of 6 analyzed; in 5 patients, staining for pSYK was increased in LN- compared with PB-derived cells (median 7-fold increase).
After in vitro BCR activation, most BCR signature genes were more strongly up-regulated in UM-CLL than M-CLL patients, most prominently OAS3, LPL, and GFI1. However, a few genes were equally or even more strongly induced in M-CLL patients, including CCL4 and EGR3 (Figure 4C). While the basis for such a differential regulation of BCR target genes is unknown, we were intrigued to find the same relative pattern of gene expression in vivo. In LN-derived cells, OAS3, LPL, and GFI1 were also more highly induced in UM-CLL patients than in M-CLL patients, and EGR3 was one of the few genes more strongly induced in M-CLL cells (Figure 4D). In summary, the gene expression response to BCR engagement was consistently stronger in UM-CLL than in M-CLL patients both in vivo and in vitro.
Expression of the CLL-BCR signature by LN-derived cells could indicate direct BCR activation in the LN, or it could be due to homing of activated cells to the LN. To analyze a more proximal event in BCR activation, we used flow cytometry to detect phosphorylation of SYK, which becomes detectable within minutes after BCR activation. In LN- but not in BM-derived cells, we detected increased phosphorylation of SYK compared with PB cells (Figure 4E). These data support antigen-dependent BCR activation in CLL cells in vivo, and suggest that in the majority of cases the LN is the site where full BCR activation occurs.
Activation of the canonical NF-κB pathway in LN-resident CLL cells
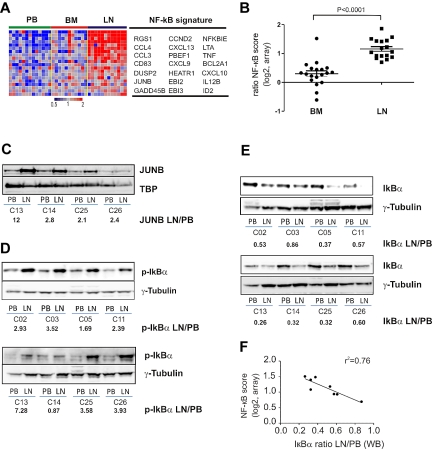
Many genes up-regulated in LN-derived cells indicated activation of the NF-κB pathway. Using GSEA, we found that 3 NF-κB signatures previously identified in B-cell lymphoma cell lines having constitutive activation of the canonical pathway23 were highly enriched in LN- compared with PB-derived cells (FDR < 0.0001; Table 2; Figure 5A). These NF-κB target genes are involved in cell-cycle regulation (CCND2), inhibition of apoptosis (BCL2A1), signal transduction (JUNB, DUSP2), and chemotaxis (CCL3, CCL4, and RGS1). We averaged the expression level of the most significantly enriched NF-κB genes to yield a “NF-κB score” (Figure 5B). Compared with the matched PB sample, all LN-derived cells showed an increase in the NF-κB score (n = 17, median 2.08-fold). In contrast, BM-derived cells in general showed only a slight increase in the NF-κB score compared with PB-derived cells (n = 19, median 1.25-fold). Confirming the gene expression changes, we found increased protein expression of JUNB in LN-resident cells (Figure 5C).
Figure 5.
NF-κB is activated through the canonical pathway in LN-derived CLL cells. (A) Genes in the NF-κB signature that were most significantly enriched in the LN as identified by GSEA (leading edge genes) are depicted in a heat map for the 12 patients having contributed cells from all 3 compartments. (B) The NF-κB score was computed as the average of the mRNA expression level of the leading edge genes for each sample. Shown is the ratio of the BCR score in BM (n = 19) or LN (n = 17) relative to the score of the matched PB sample. Comparison between BM and LN was by Student t test. The sample with the exceptionally high score in the BM was obtained from CLL_C10. (C) Western blot of nuclear lysates from purified CLL cells; JUNB expression was normalized to TBP and the ratio between LN- and PB-derived cells is shown. Western blots of cytoplasmic protein fractions from purified CLL cells were probed for phosphorylated IκBα (p-IκBα, Ser 32-36; D) and for total IκBα (E). IκBα and p-IκBα expression quantified by densitometry was normalized to γ-tubulin and the ratio between LN- and PB-derived cells is shown. (F) Pearson correlation between IκBα expression and the NF-κB gene expression score.
The NF-κB transcription factor family consists of several subunits that are regulated through 2 general pathways.24 In the canonical pathway, the inhibitor IκBα binds cytosolic p50 and p65. Upon activation, the kinase IKKβ phosphorylates IκBα, prompting its degradation and the release of p50 and p65, which then translocate to the nucleus and activate transcription. Consistent with activation of the canonical NF-κB pathway, we detected increased phosphorylation of IκBα in LN-derived tumor cells compared with the matched PB-derived cells (Figure 5D). Consequently, the level of total IκBα was reduced by > 50% in LN-derived cells (Figure 5E), and total IκBα levels were inversely correlated with the NF-κB gene expression score (r2 = −0.76; Figure 5F).
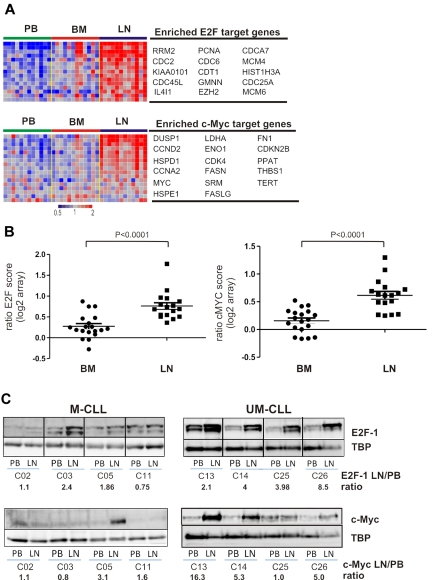
Tumor proliferation and c-MYC activation in the tissue microenvironment
Among the genes up-regulated in the LN, GSEA identified several gene sets indicating tumor proliferation. Most significant were gene sets regulated by the E2F family of transcription factors and by c-MYC (Table 2, Figure 6A). We derived a “proliferation score” by averaging the expression of the most highly enriched E2F target genes. Compared with the matched PB sample, all LN-derived cells showed an increase in proliferation (n = 17, median 1.61-fold, Figure 6B) while BM-derived cells typically showed only a slight increase (n = 19, median 1.16-fold). However, in 3 patients, BM-derived cells also showed clearly increased proliferation compared with PB cells, and in 2 of these, BM- and LN-derived cells had equal proliferation scores. A second group of proliferation-related gene sets enriched in LN-derived cells are controlled by c-MYC (Table 2, Figure 6A). c-MYC, a key transcription factor involved in lymphomagenesis and G1/S transition, cooperates with E2F1 to promote cell proliferation.25 Similar to our findings for E2F-regulated genes, c-MYC target genes were clearly up-regulated in all LN-derived cells (n = 17, median 1.53-fold, Figure 6B), but less so in BM-derived cells (n = 19, median 1.13-fold, Figure 6B). Thus, in the majority of samples, the LN microenvironment harbors the proliferative fraction of the clone. Furthermore, c-MYC and its target genes are up-regulated in LN-resident CLL cells.
Figure 6.
Tumor proliferation and c-MYC activation in the tissue microenvironment. (A) Genes of the E2F and c-MYC signatures that were most significantly enriched in the LN as identified by GSEA (leading-edge genes) are depicted in a heat map for the 12 patients having contributed cells from all 3 compartments. For E2F only genes that were at least 1.5-fold more highly expressed in LN than PB are shown. (B) The E2F and c-MYC scores were computed individually as the average of the mRNA expression level of the leading-edge genes for each sample. Shown is the ratio of the E2F and c-MYC scores in BM (n = 19) or LN (n = 17) relative to the score of the matched PB sample. Comparison between BM and LN was by Student t test. (C) Western blot of nuclear protein extracts from purified CLL cells were probed with anti-E2F1 or anti–c-MYC antibody, quantified by densitometry, and normalized to TBP. The ratios of LN- to PB-derived cells for E2F1 and c-MYC protein expression are indicated.
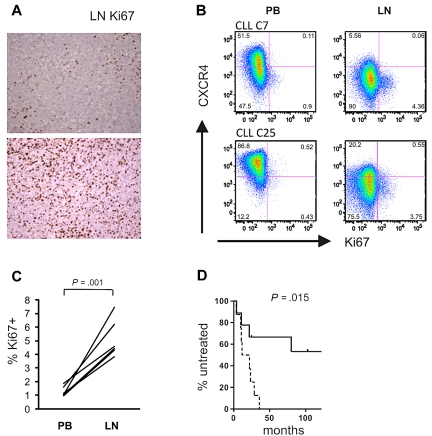
To confirm the gene expression data at the protein level, we analyzed expression of E2F1, a transcription factor that drives the expression of genes essential for G1/S phase transition, in LN- and matched PB-derived CLL cells. In 6 of 8 sample pairs analyzed, we found increased E2F1 expression in the LN sample. Notably, E2F1 up-regulation in the LN was present in all UM-CLL samples, but in only half of the M-CLL samples analyzed (Figure 6C). Western blotting also confirmed increased levels of c-MYC in LN-derived cells, again with preferential up-regulation of c-MYC in UM-CLL cells (Figure 6C). Next we assessed staining for Ki67 in LN biopsies and found cases that showed a preferential location of staining in areas identified as proliferation centers, whereas others showed more diffuse staining throughout the node (Figure 7A). We used flow cytometry to be able to compare Ki67 between LN- and PB-derived CLL cells (Figure 7B). As expected, in PB the frequency of Ki67-positive cells was low. In LN, 2%-7% of cells stained for Ki67, a median 4-fold more than in matched PB-derived CLL cells (Figure 7C).
Figure 7.
Tumor proliferation in the LN correlates with disease progression. (A) Magnification (×200) of LN biopsies of 2 representative CLL samples: top panel, preferential Ki67 staining in proliferation centers; bottom panel, diffuse Ki67 positivity. (B) Ki67 expression was assessed by flow cytometry in CD3− cells from the indicated anatomical sites. Shown are 2 representative patients of a total of 6 analyzed. Cutoffs in each sample were chosen so that PB cells were around 1% Ki67+ cells. (C) Ki67 expression in CD3− cells from the indicated anatomic sites. Statistical comparison was by paired Student t test. (D) Kaplan-Meier analysis of time to treatment in all patients with LN biopsies (n = 17) based on E2F scores: dashed line, scores above the median; solid line, scores below the median.
Finally, tumor proliferation in the LN was correlated with clinical outcome. The E2F score in LN-derived cells predicted time to treatment measured as the time from diagnosis of CLL to the development of active disease and the initiation of treatment (Figure 7D). Patients with an E2F score above the median had a median time to treatment of 16.7 months, compared with 113 months for patients with a low score (P = .015). The proliferation score in LN tended to be higher in patients with UM-CLL than in those with M-CLL (supplemental Figure 4), and all patients with an E2F score above the median were of the UM-CLL subtype.
Discussion
We discovered a pronounced effect of the tissue microenvironment on the tumor biology of CLL cells in vivo. In prior studies, gene expression profiling of peripheral blood cells characterized CLL as a malignancy of quiescent cells related to memory B cells.2,3 By comparing the gene expression of CLL cells located to different anatomic compartments, we identified the LN as a site of CLL cell activation and tumor proliferation. We found similar but less pronounced changes in CLL cells in the BM, suggesting distinct effects of the LN and BM microenvironment on the activation of signaling pathways and CLL tumor biology. One caveat, however, is that we cannot ensure equal representation of all BM-resident CLL cells in the aspirate. It is conceivable that tumor cells in close contact with the stroma or the endosteal surface may be underrepresented in the analysis, and we may therefore have underestimated the contribution of the BM microenvironment.
A key objective of this study was to identify which signaling pathways are engaged in CLL cells in vivo. Given the many factors reported to enhance CLL cell survival in vitro,13,14 we found a surprisingly short list of factors that were dominated by BCR signaling. The BCR is an essential signal transduction pathway for the survival and proliferation of mature B lymphocytes. Antigen likely plays a role at one point during the ontogeny of CLL,26 possibly in the context of positive selection of a precursor cell, as described for B1 cells in the mouse.27 In the present study, we identified inducible, probably antigen-driven BCR signaling in CLL cells in the LN that resulted in SYK activation and the induction of a characteristic gene expression signature. Remarkably, in vitro studies also identified up-regulation of BCR target genes in CLL cells cocultured on nurselike cells, including CCL3, CCL4, EGR2, EGR3, and MYC, and this effect could be blocked by the small-molecule SYK inhibitor R406.28 While we have not shown a direct link between BCR activation in the LN and disease progression, this can be reasonably inferred. Antigen-dependent BCR activation has been shown to accelerate disease progression in a mouse lymphoma model,29 and in some mucosa-associated lymphoid tissue (MALT) lymphomas, there is an antigen-dependent phase of lymphomagenesis during which antigen removal leads to regression of disease.30,31 Furthermore, EGR-1, one of the BCR target genes most highly induced in the LN, is required for proliferation in response to BCR activation and is essential for marginal zone B-cell development.32 Ongoing BCR activation in secondary lymphoid tissues thus emerges as a key pathway propagating clonal expansion in CLL.
BCR activation in the LN was more prominent in patients with UM-CLL than in those with M-CLL. This conclusion is based on the quantitative measurement of 60 validated BCR target genes. Most of these genes were also more strongly induced in UM-CLL patients after IgM cross-linking in vitro. These findings may explain the observation of a preferential expression of BCR-regulated genes in PB-derived tumor cells of the UM-CLL subtype compared with M-CLL,3 which, in light of our findings, appear as an “echo” of BCR activation in the LN. Similarly, expression of LPL, the most strongly up-regulated BCR target gene in UM-CLL in the LN, has been described as a surrogate marker for UM-CLL.33,34 After in vitro IgM ligation, UM-CLL had prolonged SYK activation and stronger induction of BCR target genes, consistent with observations that ZAP-70 expression can enhance and prolong the BCR signal.8,10 In addition, different antigen specificities of UM-CLL and M-CLL cells may affect the frequency and strength of BCR activation. UM-CLL cells that express a more polyreactive BCR35 may have more frequent antigen contact. A role for 2 factors, 1 intrinsic (ZAP-70) and 1 extrinsic (antigen), in determining BCR signal strength could explain clinical differences in disease progression between different CLL subtypes. ZAP-70–positive CLL progresses rapidly irrespective of the IGHV mutation status, whereas among ZAP-70–negative patients, UM-CLL progresses more rapidly than M-CLL.36
BCR engagement results in the activation of downstream signaling pathways, including NF-κB and calcineurin-NFAT, both of which are up-regulated in LN-resident CLL cells (Table 2). Studies on NF-κB signaling in CLL cells in vivo have been limited to the analysis of PB cells and reported variable but overall increased NF-κB activity in CLL cells compared with circulating naive B cells.37 Furthermore, expression of p65 by leukemic cells has been suggested as a predictor of clinical outcome.38 In contrast to those studies viewing NF-κB as constitutively active in CLL, our data indicate that tumor cells acquire NF-κB activation through specific interactions in the microenvironment. Cells exiting the microenvironment presumably maintain NF-κB activation for some time, suggesting that the reported expression of p65 in circulating cells could reflect the strength of NF-κB induction in the microenvironment.
In addition to the BCR, GSEA indicated activation of tumor necrosis factor and Toll receptor pathways that could contribute to NF-κB activation in the LN. A role for tumor necrosis factor family members, especially BAFF (B-cell activating factor),39 in CLL cell survival has been suggested by in vitro studies, and there are early indications that TLR signaling could be a survival pathway in CLL as well.40 TLR ligands such as bacterial cell membrane components or unmethylated DNA are potent stimulators of immune cells, activate the NF-κB pathway, and may cooperate with BCR signals to overcome the anergy of autoreactive B cells.41 The relative contribution of these pathways to NF-κB activation in CLL cells in vivo remains to be defined.
CLL has been regarded as an accumulative disorder of lymphocytes with a defect in apoptosis. This view was largely based on the analysis of PB cells, which are arrested in the G0/G1 phase of the cell cycle. In contrast, using deuterated water to label proliferating cells in vivo, Messmer et al identified a fraction of the CLL clone, ranging from 0.1%-1.76%, that is generated each day.12 In the present study, we identify the LN as an important site of tumor proliferation, while cells in the BM generally had lower proliferation scores. Consistent with the more rapid clinical progression of the UM subtype, we also found that UM-CLL cells had higher proliferation rates than M-CLL cells. Our observations support the emerging notion that proliferation above apoptosis resistance determines clinical outcome in CLL.
The acquisition of additional genetic lesions in the leukemic clone greatly influences clinical outcome.42 For unclear reasons, UM-CLL patients acquire cytogenetic abnormalities that are associated with adverse outcomes more frequently and/or more rapidly than M-CLL patients.43 We identified possible factors that could contribute to the genomic instability in UM-CLL: increased proliferation and higher c-MYC expression.44 An important factor in genomic instability is telomere shortening, which has been reported to be more significant in UM-CLL than in M-CLL.45 Increased proliferation in UM-CLL leading to telomere shortening could contribute to clonal evolution by selecting for cells with a defective DNA damage response.46 In keeping with accentuated genotoxic stress in proliferating cells, p53 gene expression signatures were significantly enriched in LN-derived cells (Table 2).
How can these data inform the treatment of CLL patients? Current chemotherapy options induce extended remissions but presumably no cure. Relapsed disease is characterized by the presence of additional cytogenetic abnormalities and relatively inferior response to subsequent treatment.47 Our findings that tumor proliferation is a result of tumor-host interactions in vivo and is a key determinant of clinical outcome can redirect therapeutic efforts. First, inhibiting the trafficking of CLL cells to protective microenvironment niches may deprive the tumor cells of essential signals and induce apoptosis or sensitize the tumor to the effect of cytotoxic therapy.48 Furthermore, targeting essential signaling pathways in the microenvironment with small molecules could inhibit proliferation of CLL cells, and thereby not only shrink tumor burden but also reduce clonal evolution. We identify the BCR as a valid therapeutic target in vivo, and small molecules inhibiting BCR signaling indeed show promise in early clinical trials.49 In addition, if distinct antigens drive CLL progression, then strategies to inhibit antigen binding to the BCR could be developed.35 Such strategies, which are devoid of the side effects of conventional chemotherapy, could be applied early in the clinical course to prevent disease progression. Another implication of our study is that tumor biology differs between anatomic locations, possibly affecting drug sensitivity. For example, we found increased expression of BCL2A1, an NF-κB regulated gene in LN-resident cells. BCL2A1 is an antiapoptotic member of the BCL-2 family of proteins that can confer resistance to BCL-2 inhibitors.50 Thus, the variable biology of CLL cells in the microenvironment presents therapeutic opportunities and challenges that will have to be addressed in carefully designed clinical trials.
Supplementary Material
Acknowledgments
Foremost we thank our patients for their willingness to undergo additional or more extensive procedures to make this research possible. We thank Dr Lou Staudt for lymphochip microarrays, the NCI surgery team for performing excisional lymph node biopsies, and Theresa Davies-Hill for assistance in processing biopsy specimens. We thank Drs Lou Staudt, Neal Young, Cindy Dunbar, and Clifton Mo for helpful discussions and critical reading of the manuscript. We gratefully acknowledge the American Physician Fellowship (APF) for support of Y.H.
This research was also supported by the Intramural Research Program of the NIH, the National Heart, Lung, and Blood Institute, and the National Cancer Institute.
Footnotes
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: Y.H., P.P.-G., and A.W. designed the study, analyzed data, and wrote the paper; Y.H., B.V., F.G., N.N., E.L., and L.S. conducted molecular and cellular assays and performed data analysis; D.L. and P.M. conducted statistical analysis; A.B., J.P.M., M.S.-S., and C.Y. performed and analyzed flow cytometry; N.R. and P.L. performed gene expression profiling; D.C.A. conducted fluorescence in situ hybridization analysis; S.P., I.M., and M.R. provided pathology evaluations; R.S. coordinated and performed surgical procedures; W.H.W. initiated and supervised the clinical trial; T.W., G.E.M., W.H.W., and A.W. conducted the clinical trial and evaluated the patients; and A.W. coordinated and supervised the laboratory studies.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Adrian Wiestner, MD, PhD, Hematology Branch, National Heart, Lung, and Blood Institute, NIH, Bldg 10, CRC 3-5140, 10 Center Dr, Bethesda, MD 20892-1202; e-mail: wiestnera@mail.nih.gov.
References
- 1.Chiorazzi N, Rai KR, Ferrarini M. Chronic lymphocytic leukemia. N Engl J Med. 2005;352(8):804–815. doi: 10.1056/NEJMra041720. [DOI] [PubMed] [Google Scholar]
- 2.Klein U, Tu Y, Stolovitzky GA, et al. Gene expression profiling of B cell chronic lymphocytic leukemia reveals a homogeneous phenotype related to memory B cells. J Exp Med. 2001;194(11):1625–1638. doi: 10.1084/jem.194.11.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rosenwald A, Alizadeh AA, Widhopf G, et al. Relation of gene expression phenotype to immunoglobulin mutation genotype in B cell chronic lymphocytic leukemia. J Exp Med. 2001;194(11):1639–1647. doi: 10.1084/jem.194.11.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wiestner A, Rosenwald A, Barry TS, et al. ZAP-70 expression identifies a chronic lymphocytic leukemia subtype with unmutated immunoglobulin genes, inferior clinical outcome, and distinct gene expression profile. Blood. 2003;101(12):4944–4951. doi: 10.1182/blood-2002-10-3306. [DOI] [PubMed] [Google Scholar]
- 5.Crespo M, Bosch F, Villamor N, et al. ZAP-70 expression as a surrogate for immunoglobulin-variable-region mutations in chronic lymphocytic leukemia. N Engl J Med. 2003;348(18):1764–1775. doi: 10.1056/NEJMoa023143. [DOI] [PubMed] [Google Scholar]
- 6.Orchard JA, Ibbotson RE, Davis Z, et al. ZAP-70 expression and prognosis in chronic lymphocytic leukaemia. Lancet. 2004;363(9403):105–111. doi: 10.1016/S0140-6736(03)15260-9. [DOI] [PubMed] [Google Scholar]
- 7.Rassenti LZ, Huynh L, Toy TL, et al. ZAP-70 compared with immunoglobulin heavy-chain gene mutation status as a predictor of disease progression in chronic lymphocytic leukemia. N Engl J Med. 2004;351(9):893–901. doi: 10.1056/NEJMoa040857. [DOI] [PubMed] [Google Scholar]
- 8.Chen L, Huynh L, Apgar J, et al. ZAP-70 enhances IgM signaling independent of its kinase activity in chronic lymphocytic leukemia. Blood. 2008;111(5):2685–2692. doi: 10.1182/blood-2006-12-062265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cutrona G, Colombo M, Matis S, et al. Clonal heterogeneity in chronic lymphocytic leukemia cells: superior response to surface IgM cross-linking in CD38, ZAP-70-positive cells. Haematologica. 2008;93(3):413–422. doi: 10.3324/haematol.11646. [DOI] [PubMed] [Google Scholar]
- 10.Gobessi S, Laurenti L, Longo PG, Sica S, Leone G, Efremov DG. ZAP-70 enhances B-cell-receptor signaling despite absent or inefficient tyrosine kinase activation in chronic lymphocytic leukemia and lymphoma B cells. Blood. 2007;109(5):2032–2039. doi: 10.1182/blood-2006-03-011759. [DOI] [PubMed] [Google Scholar]
- 11.Richardson SJ, Matthews C, Catherwood MA, et al. ZAP-70 expression is associated with enhanced ability to respond to migratory and survival signals in B-cell chronic lymphocytic leukemia (B-CLL). Blood. 2006;107(9):3584–3592. doi: 10.1182/blood-2005-04-1718. [DOI] [PubMed] [Google Scholar]
- 12.Messmer BT, Messmer D, Allen SL, et al. In vivo measurements document the dynamic cellular kinetics of chronic lymphocytic leukemia B cells. J Clin Invest. 2005;115(3):755–764. doi: 10.1172/JCI23409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burger JA, Ghia P, Rosenwald A, Caligaris-Cappio F. The microenvironment in mature B-cell malignancies: a target for new treatment strategies. Blood. 2009;114(16):3367–3375. doi: 10.1182/blood-2009-06-225326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deaglio S, Malavasi F. Chronic lymphocytic leukemia microenvironment: shifting the balance from apoptosis to proliferation. Haematologica. 2009;94(6):752–756. doi: 10.3324/haematol.2009.006676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davis RE, Ngo VN, Lenz G, et al. Chronic active B-cell-receptor signalling in diffuse large B-cell lymphoma. Nature. 2010;463(7277):88–92. doi: 10.1038/nature08638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guarini A, Chiaretti S, Tavolaro S, et al. BCR ligation induced by IgM stimulation results in gene expression and functional changes only in IgV H unmutated chronic lymphocytic leukemia (CLL) cells. Blood. 2008;112(3):782–792. doi: 10.1182/blood-2007-12-127688. [DOI] [PubMed] [Google Scholar]
- 17.Ghia P, Chiorazzi N, Stamatopoulos K. Microenvironmental influences in chronic lymphocytic leukaemia: the role of antigen stimulation. J Intern Med. 2008;264(6):549–562. doi: 10.1111/j.1365-2796.2008.02030.x. [DOI] [PubMed] [Google Scholar]
- 18.Shaffer AL, Wright G, Yang L, et al. A library of gene expression signatures to illuminate normal and pathological lymphoid biology. Immunol Rev. 2006;210:67–85. doi: 10.1111/j.0105-2896.2006.00373.x. [DOI] [PubMed] [Google Scholar]
- 19.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alizadeh AA, Eisen MB, Davis RE, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 21.Burger JA, Burger M, Kipps TJ. Chronic lymphocytic leukemia B cells express functional CXCR4 chemokine receptors that mediate spontaneous migration beneath bone marrow stromal cells. Blood. 1999;94(11):3658–3667. [PubMed] [Google Scholar]
- 22.Vallat LD, Park Y, Li C, Gribben JG. Temporal genetic program following B-cell receptor cross-linking: altered balance between proliferation and death in healthy and malignant B cells. Blood. 2007;109(9):3989–3997. doi: 10.1182/blood-2006-09-045377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lam LT, Davis RE, Pierce J, et al. Small molecule inhibitors of IkappaB kinase are selectively toxic for subgroups of diffuse large B-cell lymphoma defined by gene expression profiling. Clin Cancer Res. 2005;11(1):28–40. [PubMed] [Google Scholar]
- 24.Hayden MS, Ghosh S. Shared principles in NF-kappaB signaling. Cell. 2008;132(3):344–362. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 25.Baudino TA, Maclean KH, Brennan J, et al. Myc-mediated proliferation and lymphomagenesis, but not apoptosis, are compromised by E2f1 loss. Mol Cell. 2003;11(4):905–914. doi: 10.1016/s1097-2765(03)00102-3. [DOI] [PubMed] [Google Scholar]
- 26.Chiorazzi N, Ferrarini M. B cell chronic lymphocytic leukemia: lessons learned from studies of the B cell antigen receptor. Annu Rev Immunol. 2003;21:841–894. doi: 10.1146/annurev.immunol.21.120601.141018. [DOI] [PubMed] [Google Scholar]
- 27.Hayakawa K, Asano M, Shinton SA, et al. Positive selection of natural autoreactive B cells. Science. 1999;285(5424):113–116. doi: 10.1126/science.285.5424.113. [DOI] [PubMed] [Google Scholar]
- 28.Burger JA, Quiroga MP, Hartmann E, et al. High-level expression of the T-cell chemokines CCL3 and CCL4 by chronic lymphocytic leukemia B cells in nurselike cell cocultures and after BCR stimulation. Blood. 2009;113(13):3050–3058. doi: 10.1182/blood-2008-07-170415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Refaeli Y, Young RM, Turner BC, Duda J, Field KA, Bishop JM. The B cell antigen receptor and overexpression of MYC can cooperate in the genesis of B cell lymphomas. PLoS Biol. 2008;6(6):e152. doi: 10.1371/journal.pbio.0060152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hermine O, Lefrere F, Bronowicki JP, et al. Regression of splenic lymphoma with villous lymphocytes after treatment of hepatitis C virus infection. N Engl J Med. 2002;347(2):89–94. doi: 10.1056/NEJMoa013376. [DOI] [PubMed] [Google Scholar]
- 31.Wotherspoon AC, Doglioni C, Diss TC, et al. Regression of primary low-grade B-cell gastric lymphoma of mucosa- associated lymphoid tissue type after eradication of Helicobacter pylori. Lancet. 1993;342(8871):575–577. doi: 10.1016/0140-6736(93)91409-f. [DOI] [PubMed] [Google Scholar]
- 32.Gururajan M, Simmons A, Dasu T, et al. Early growth response genes regulate B cell development, proliferation, and immune response. J Immunol. 2008;181(7):4590–4602. doi: 10.4049/jimmunol.181.7.4590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heintel D, Kienle D, Shehata M, et al. High expression of lipoprotein lipase in poor risk B-cell chronic lymphocytic leukemia. Leukemia. 2005;19(7):1216–1223. doi: 10.1038/sj.leu.2403748. [DOI] [PubMed] [Google Scholar]
- 34.Oppezzo P, Vasconcelos Y, Settegrana C, et al. The LPL/ADAM29 expression ratio is a novel prognosis indicator in chronic lymphocytic leukemia. Blood. 2005;106(2):650–657. doi: 10.1182/blood-2004-08-3344. [DOI] [PubMed] [Google Scholar]
- 35.Seiler T, Woelfle M, Yancopoulos S, et al. Characterization of structurally defined epitopes recognized by monoclonal antibodies produced by chronic lymphocytic leukemia B cells. Blood. 2009;114(17):3615–3624. doi: 10.1182/blood-2009-01-197822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rassenti LZ, Jain S, Keating MJ, et al. Relative value of ZAP-70, CD38, and immunoglobulin mutation status in predicting aggressive disease in chronic lymphocytic leukemia. Blood. 2008;112(5):1923–1930. doi: 10.1182/blood-2007-05-092882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hewamana S, Alghazal S, Lin TT, et al. The NF-kappaB subunit Rel A is associated with in vitro survival and clinical disease progression in chronic lymphocytic leukemia and represents a promising therapeutic target. Blood. 2008;111(9):4681–4689. doi: 10.1182/blood-2007-11-125278. [DOI] [PubMed] [Google Scholar]
- 38.Hewamana S, Lin TT, Rowntree C, et al. Rel a is an independent biomarker of clinical outcome in chronic lymphocytic leukemia. J Clin Oncol. 2009;27(5):763–769. doi: 10.1200/JCO.2008.19.1114. [DOI] [PubMed] [Google Scholar]
- 39.Endo T, Nishio M, Enzler T, et al. BAFF and APRIL support chronic lymphocytic leukemia B-cell survival through activation of the canonical NF-kappaB pathway. Blood. 2007;109(2):703–710. doi: 10.1182/blood-2007-04-081786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muzio M, Bertilaccio MT, Simonetti G, Frenquelli M, Caligaris-Cappio F. The role of Toll-like receptors in chronic B cell malignancies. Leuk Lymphoma. 2009;50(10):1573–1580. doi: 10.1080/10428190903115410. [DOI] [PubMed] [Google Scholar]
- 41.Leadbetter EA, Rifkin IR, Hohlbaum AM, Beaudette BC, Shlomchik MJ, Marshak-Rothstein A. Chromatin-IgG complexes activate B cells by dual engagement of IgM and Toll-like receptors. Nature. 2002;416(6881):603–607. doi: 10.1038/416603a. [DOI] [PubMed] [Google Scholar]
- 42.Döhner H, Stilgenbauer S, Benner A, et al. Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med. 2000;343(26):1910–1916. doi: 10.1056/NEJM200012283432602. [DOI] [PubMed] [Google Scholar]
- 43.Roos G, Krober A, Grabowski P, et al. Short telomeres are associated with genetic complexity, high-risk genomic aberrations, and short survival in chronic lymphocytic leukemia. Blood. 2008;111(4):2246–2252. doi: 10.1182/blood-2007-05-092759. [DOI] [PubMed] [Google Scholar]
- 44.Felsher DW, Bishop JM. Transient excess of MYC activity can elicit genomic instability and tumorigenesis. Proc Natl Acad Sci U S A. 1999;96(7):3940–3944. doi: 10.1073/pnas.96.7.3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Damle RN, Batliwalla FM, Ghiotto F, et al. Telomere length and telomerase activity delineate distinctive replicative features of the B-CLL subgroups defined by immunoglobulin V gene mutations. Blood. 2004;103(2):375–382. doi: 10.1182/blood-2003-04-1345. [DOI] [PubMed] [Google Scholar]
- 46.Calado RT, Regal JA, Hills M, et al. Constitutional hypomorphic telomerase mutations in patients with acute myeloid leukemia. Proc Natl Acad Sci U S A. 2009;106(4):1187–1192. doi: 10.1073/pnas.0807057106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tam CS, Wierda W, O'Brien S, et al. Life after fludarabine, cylcophosphamide, & rituximab (FCR)–the clinical outcome of patients with chronic lymphocytic leukemia who receive salvage treatment after frontline FCR [abstract]. Blood. 2008;(112):2090. [Google Scholar]
- 48.Burger JA, Peled A. CXCR4 antagonists: targeting the microenvironment in leukemia and other cancers. Leukemia. 2009;23(1):43–52. doi: 10.1038/leu.2008.299. [DOI] [PubMed] [Google Scholar]
- 49.Friedberg JW, Sharman J, Sweetenham J, et al. Inhibition of Syk with fostamatinib disodium has significant clinical activity in non-Hodgkin lymphoma and chronic lymphocytic leukemia. Blood. 2010;115(13):2578–2585. doi: 10.1182/blood-2009-08-236471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vogler M, Butterworth M, Majid A, et al. Concurrent up-regulation of BCL-XL and BCL2A1 induces approximately 1000-fold resistance to ABT-737 in chronic lymphocytic leukemia. Blood. 2009;113(18):4403–4413. doi: 10.1182/blood-2008-08-173310. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.