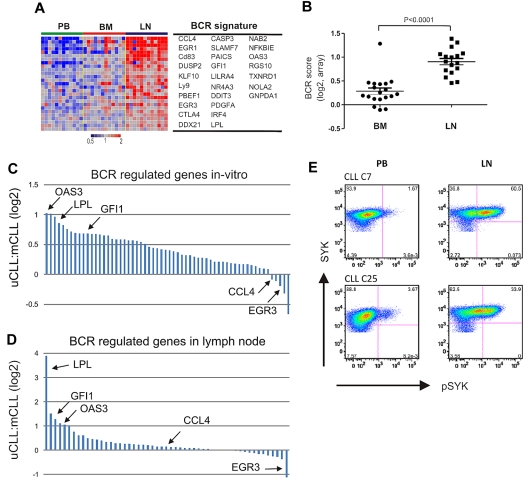
Figure 4.
BCR activation in LN-resident CLL cells. (A) Genes in the CLL-BCR signature that were most significantly enriched in the LN as identified by GSEA (leading-edge genes) are depicted in a heat map for the 12 patients having contributed cells from all 3 compartments. (B) The BCR score was computed as the average of the mRNA expression level of the leading edge genes for each sample. Shown is the ratio of the BCR score in BM (n = 19) or LN (n = 17) relative to the score of the matched PB sample. Comparison between BM and LN was by Student t test. The sample with exceptionally high score in BM was obtained from CLL_C10. (C, D) Relative gene expression changes of BCR-regulated genes between UM-CLL and M-CLL. Each gene is represented by a bar, select genes are highlighted. (C) Ratio of relative gene expression change after IgM cross-linking between UM-CLL (n = 4) and M-CLL (n = 4). Bars represent 61 genes, as described in Figure 3A. The fold up-regulation of each gene was averaged for each subtype and the ratio between UM-CLL and M-CLL is shown on a log2 scale. (D) Ratio of relative gene expression change in UM-CLL (n = 12) and M-CLL (n = 5) between the LN- and matched PB-derived cells. Bars represent all 61 genes as in panel C. The fold up-regulation of each gene in the LN-derived cells compared with the matched PB sample was averaged within each subtype, and the ratio between UM-CLL and M-CLL is shown on a log2 scale. (E) Phosphorylation of SYK was assessed by flow cytometry in CD3− cells from the indicated anatomical site. Shown are 2 representative patients of a total of 6 analyzed; in 5 patients, staining for pSYK was increased in LN- compared with PB-derived cells (median 7-fold increase).