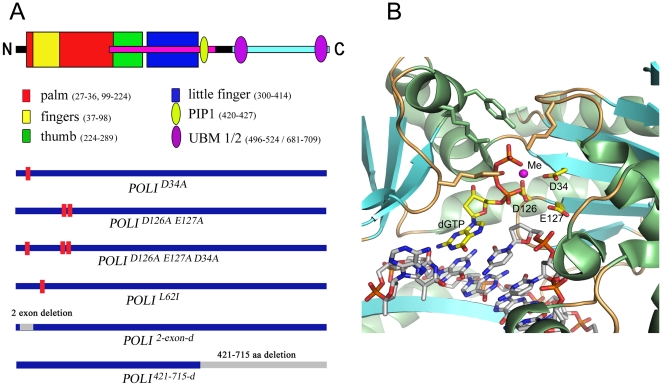
Figure 1. Structure of Pol ι and the variants studied.
A. Upper half. The schematic domain structure of Pol ι is shown (PIP is the protein interaction domain, UBM – ubiqutin-binding motif). Lower half. The Pol ι mutant variants were used in the study (Red bars on the thick blue line representing Pol ι show the positions of amino acid changes in polymorphic variants of Pol ι. Grey bars indicate deletions for truncated Pol ι variants): hPOLISc2exon-d (Pol ι 2exon-d) – Pol ι variant with a deletion of exon 2 (encoding for amino acids 14-55) representing an alternative splice variant of human and mouse Pol ι; hPOLIScD34A (Pol ι D34A) and hPOLIScD126A/E127A (Pol ι D126A/E127A) – “catalytically dead” Pol ι variants created as amino acid substitutions of evolutionary conservative Asp34 or Asp126 and Glu127 to Ala; hPOLIScD34A/D126A/E127A (Pol ι D34A/D126A/E127A) – a triple “catalytically dead” Pol ι variant with amino acid substitutions of Asp34, Asp126 and Glu127 to Ala; hPOLIScL62I (Pol ιL62I) – Pol ι variant with a substitution of evolutionary polymorphic amino acid Leu62 to Ile; hPOLISc42I-612-d variant with a deletion of the C-terminal half of Pol ι is shown to illustrate what minimal part retains enzymatic activity and whose crystal structure has been determined. B. Polι active site. A close view at the Pol ι active site in ternary complex with DNA (template T) and incoming dGTP (3gv8). Pol ι and DNA molecules are represented as cartoon and sticks, respectively. The incorporated nucleotides and active site residues Asp34, Asp126 and Glu127 are represented by sticks and highlighted with yellow carbons. The side chains of phosphate-binding residues are also shown as sticks. The metal ion is drawn as a magenta ball.