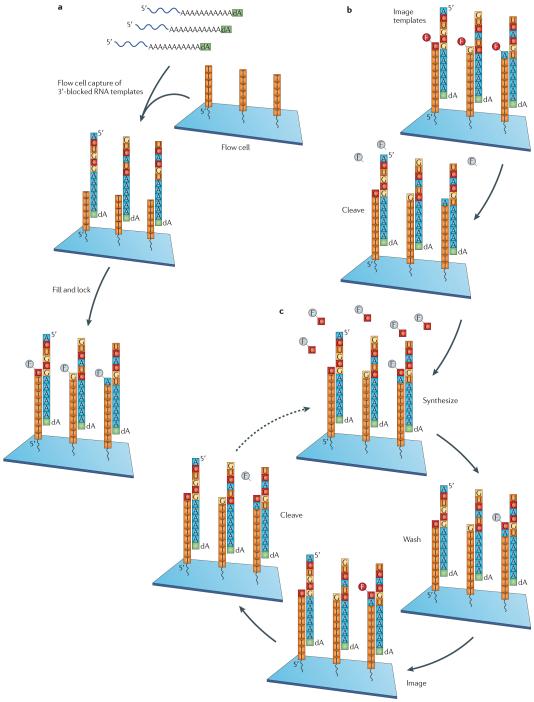
Figure 4. Direct RNA sequencing using the Helicos approach.
a | RNA that is polyadenylated and 3′ deoxy-blocked with poly(A) polymerase is captured on poly(dT)-coated surfaces. A ‘fill-and-lock’ step is performed, in which the ‘fill’ step is performed with natural thymidine and polymerase, and the ‘lock’ step is performed with fluorescently labelled A, C and G Virtual Terminator (VT) nucleotides104 and polymerase. This step corrects for any misalignments that may be present in poly(A) and poly(T) duplexes, and ensures that the sequencing starts in the RNA template rather than the polyadenylated tail. b | Imaging is performed to locate the positions of the templates. Then, chemical cleavage of the dye–nucleotide linker is performed to release the dye and prepare the templates for nucleotide incorporation. c | Incubation of this surface with one labelled nucleotide (C-VT is shown as an example) and a polymerase mixture is carried out. After this step, imaging is performed to locate the templates that have incorporated the nucleotide. Chemical cleavage of the dye allows the surface and DNA templates to be ready for the next nucleotide-addition cycle. Nucleotides are added in the C, T, A, G order for 120 total cycles (30 additions of each nucleotide).