Table 1.
Structural analysis of P450cam conformers.
| Substratea | PDB | PC1b | PC2b | F helix shiftc (Å, deg) | G helix shiftc (Å, deg) | FG helix angle (deg) | Axial Water | Missing B′ helix | Ref |
|---|---|---|---|---|---|---|---|---|---|
| P450CAM-C | |||||||||
| Camphor |
2CPP | 25.7 | 2.9 | — | — | −148.9 | No | — | (4) |
| 1YRC | 29.4 | 4.9 | 0.28, 2.7 | 0.25, 2.8 | −149.4 | (42) | |||
| 5CP4 | 24.8 | 3.7 | 0.22, 2.7 | 0.40, 1.8 | −150.1 | (43) | |||
| 2ZAX | 28.7 | 5.7 | 0.23, 3.1 | 0.28, 2.5 | −148.7 | (44) | |||
| 1DZ4 | 26.7 | 6.5 | 0.11, 1.2 | 0.55, 0.9 | −150.1 | (45) | |||
| 1S-camphor |
1AKD | 23.9 | 6.1 | 0.41, 2.7 | 0.48, 1.4 | −149.0 | No | — | (46) |
| Imidazole |
2H7Q | 24.1 | 4.9 | 0.26, 2.2 | 0.54, 1.5 | −151.4 | No | — | (47) |
Nicotine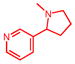
|
1P2Y | 25.9 | 5.6 | 0.08, 5.2 | 0.36, 0.5 | −148.2 | No | — | (48) |
Metyrapone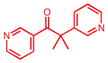
|
1PHG | 25.6 | 3.2 | 0.07, 1.4 | 0.12, 0.5 | −148.6 | No | — | (49) |
| P450CAM-I | |||||||||
| AdaC1-C8-Dans |
1RE9 | 6.4 | −9.9 | 3.30, 17.8 | 1.00, 5.9 | −156.6 | No | — | (66) |
| 1LWL | 4.4 | −11.5 | 3.59, 17.4 | 1.33, 5.4 | −156.5 | (32) | |||
| 3P6M | 2.7 | −11.2 | 3.77, 15.5 | 1.52, 5.4 | −156.7 | This study | |||
| 3P6N | 2.2 | −11.1 | 3.75, 16.0 | 1.50, 6.2 | −157.2 | This study | |||
| AdaC1-C8EtgGlu-Bio |
3OIA | 3.0 | −11.6 | 3.71, 15.8 | 1.27, 7.3 | −153.2 | No | — | Unpublished |
| AdaC1-Etg-Dans |
3P6O | 3.7 | −10.6 | 3.73, 16.7 | 1.42, 6.0 | −153.5 | No | — | This study |
| AdaC1-C6-Bio |
3P6P | 2.5 | −10.3 | 3.91, 15.2 | 1.37, 6.6 | −157.8 | No | — | This study |
| 3OH-AdaC1-C8-Dans |
3OL5 | 1.1 | −10.5 | 3.88, 14.9 | 1.96, 5.1 | −155.8 | Yes | — | Unpublished |
| P450CAM-O | |||||||||
| AdaC1-C4-Dans |
1RF9 | −9.9 | 0.7 | 3.94, 15.9 | 3.68, 10.8 | −149.7 | Yes | — | (66) |
| AdaC2-Etg-Boc |
3P6Q | −15.6 | 3.2 | 4.51, 17.2 | 3.95, 15.2 | — | Yes | 92–95 | This study |
3OH-AdaC1-Etg-Boc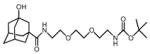
|
3P6R | −16.3 | 4.1 | 4.84, 16.6 | 4.57, 15.2 | −151.1 | No | 90–96 | This study |
| AdaC2-C8-Dansd |
3P6S | −17.3 | 2.3 | 4.59, 17.1 | 4.46, 14.9 | −150.2 | No | — | This study |
| 3P6T | −15.5 | 0.7 | 4.24, 16.8 | 4.28, 15.7 | −150.6 | This study | |||
| AdaC3-C6-Dans |
3P6U | −21.0 | 3.8 | 4.95, 17.8 | 4.83, 17.4 | −150.6 | Yes | 93–94 | This study |
3Et-AdaC1-Etg-Boc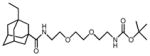
|
3P6V | −20.4 | 4.6 | 5.06, 17.4 | 5.02, 16.0 | −150.7 | No | 90–96 | This study |
| AdaC3-Etg-Boc |
3P6W | −20.5 | 3.8 | 5.04, 17.1 | 4.85, 17.4 | −151.0 | No | 91–95 | This study |
| AdaC3-C8-Dans |
3P6X | −22.2 | 3.9 | 5.07, 18.7 | 4.86, 18.1 | −150.4 | Yes | 90–94 | This study |
AdaC1-perfluorobiphenyl-Ru(bipy)3
|
1K2O | −22.7 | 3.8 | 5.54, 21.0 | 5.14, 18.8 | −151.6 | Yes | — | (65) |
| AdaC1-C8-Ru(bipy)3 |
1QMQ | −16.0 | 3.3 | 4.43, 16.0 | 4.28, 14.7 | −149.0 | Yes | — | (64) |
| Substrate-free | 3L61 | −24.9 | 4.3 | 5.27, 19.3 | 5.09, 17.9 | −150.2 | Yes | 90–97 | (18) |
| 3L62 | −21.5 | 3.6 | 4.93, 18.2 | 4.73, 16.9 | −150.2 | 91–94 | (18) | ||
Well ordered electron density included for portions of the model shown in red.
PC1 and PC2 are first two principal components in the PCA of crystal structures described in this table.
Helix shift is calculated relative to the camphor-bound closed conformation (PDB entry 2CPP) at the center of the F helix (Lys178) and G helix (Lys197).
The entire substrate was only resolved in the structure of PDB entry 3P6T.
