Abstract
Hydroxypyruvate (HP) is an intermediate of the photorespiratory pathway that originates in the oxygenase activity of the key enzyme of photosynthetic CO2 assimilation, Rubisco. In course of this high-throughput pathway, a peroxisomal transamination reaction converts serine to HP, most of which is subsequently reduced to glycerate by the NADH-dependent peroxisomal enzyme HP reductase (HPR1). In addition, a NADPH-dependent cytosolic HPR2 provides an efficient extraperoxisomal bypass. The combined deletion of these two enzymes, however, does not result in a fully lethal photorespiratory phenotype, indicating even more redundancy in the photorespiratory HP-into-glycerate conversion. Here, we report on a third enzyme, HPR3 (At1g12550), in Arabidopsis (Arabidopsis thaliana), which also reduces HP to glycerate and shows even more activity with glyoxylate, a more upstream intermediate of the photorespiratory cycle. The deletion of HPR3 by T-DNA insertion mutagenesis results in slightly altered leaf concentrations of the photorespiratory intermediates HP, glycerate, and glycine, indicating a disrupted photorespiratory flux, but not in visible alteration of the phenotype. On the other hand, the combined deletion of HPR1, HPR2, and HPR3 causes increased growth retardation, decreased photochemical efficiency, and reduced oxygen-dependent gas exchange in comparison with the hpr1xhpr2 double mutant. Since in silico analysis and proteomic studies from other groups indicate targeting of HPR3 to the chloroplast, this enzyme could provide a compensatory bypass for the reduction of HP and glyoxylate within this compartment.
Photorespiration is a high-throughput pathway that is essential for the normal growth and development of plants in air (for review, see Bauwe et al., 2010). This process is caused by Rubisco, which cannot fully discriminate between carbon dioxide and oxygen. Carboxylation of ribulose 1,5-bisphosphate by Rubisco yields two molecules, 3-phosphoglycerate (3PGA), which can directly enter the Calvin cycle. In contrast, the oxygenase activity of Rubisco produces one molecule each of 3PGA and 2-phosphoglycolate (2PG). The latter compound inhibits several essential enzymes, such as triosephosphate isomerase and phosphofructokinase (Anderson, 1971; Kelly and Latzko, 1976), and is toxic for plant cells. The photorespiratory pathway recovers and detoxifies 2PG by converting it back into the Calvin cycle intermediate, 3PGA. This requires the interplay of several enzymes located in the chloroplast, the peroxisome, the mitochondrion, and the cytosol.
One of the key intermediates of the photorespiratory cycle is hydroxypyruvate (HP), which is produced in the peroxisome from Ser by the enzyme Ala:glyoxylate (Glx) aminotransferase (AGT1; Liepman and Olsen, 2001). HP then is mostly reduced by the peroxisomal NADH-dependent HP reductase (HPR1) to d-glycerate and to a lesser extent by a cytosolic NADPH-dependent HPR2 (Timm et al., 2008). The resulting glycerate is phosphorylated to 3PGA by the chloroplastidial enzyme glycerate 3-kinase (Boldt et al., 2005) and reenters the Calvin cycle.
Deletion of photorespiratory enzymes typically leads to a strong air sensitivity of the respective mutants, which, however, can be fully recovered by elevated-CO2 conditions. While this is a distinctive feature of most photorespiratory mutants (Igarashi et al., 2003; Boldt et al., 2005; Voll et al., 2006; Schwarte and Bauwe, 2007), Arabidopsis (Arabidopsis thaliana) HPR1 knockout mutants grow nearly normally in ambient air with moderate photoperiods and show only minor changes in photosynthetic and metabolic parameters under these conditions. The additional deletion of the cytosolic HPR2 distinctly elevates the oxygen sensitivity, but this hpr1xhpr2 double mutant can still survive long-term exposure to ambient air (Timm et al., 2008). Since this indicates even more redundancy in the photorespiratory HP-into-glycerate conversion step, although with a low apparent capacity, we searched the Arabidopsis genome (Arabidopsis Genome Initiative, 2000) for related enzymes and identified protein At1g12550 as another putative HPR. Studies with the recombinant protein, which shows up to approximately 50% sequence identity with HPR1 and HPR2 and is annotated as a putative d-isomer-specific 2-hydroxyacid dehydrogenase, confirmed that At1g12550 is able to reduce HP and Glx in vitro. A physiological characterization of knockout mutants of this protein revealed minor but distinct changes in the photorespiratory cycle, as evidenced by slightly increased levels of pathway intermediates. This notwithstanding, the fact that the knockout had no effect on growth in normal air indicates that in the presence of HPR1 and HPR2, this protein plays only a minor role in the photorespiratory process. The combined deletion of HPR1, HPR2, and the newly identified HPR-like enzyme (hereafter referred to as HPR3) leads to strong impairment of growth in normal air and a massive air sensitivity after transfer from nonphotorespiratory conditions to air. Moreover, gas-exchange parameters are strongly affected in the isolated triple mutant, and we found further increased levels of Ser and related metabolites of this photorespiratory amino acid. Since HPR3 has been reported to be located in chloroplasts both by the in silico prediction database Aramemnon (Schwacke et al., 2003) and proteomic studies (Yu et al., 2008), our results provide evidence that HPR3 works as an HPR in plants and likely represents an additional bypass both to known HPRs and to glyoxylate reductases in chloroplasts.
RESULTS
Identification of a d-Isomer-Specific 2-Hydroxyacid Dehydrogenase as an Alternative HPR
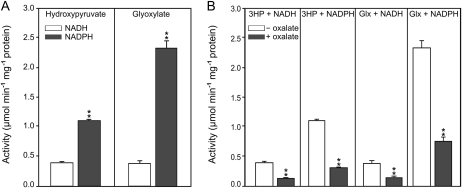
The nonlethal phenotype of the hpr1xhpr2 double mutant in ambient air indicated that additional HP-reducing enzymes could be present in Arabidopsis. For this reason, we performed a BLAST search to identify potential candidates for such enzymes. These analyses led us to a protein that is encoded by the gene At1g12550 and shares 45% sequence identity with the cytosolic HPR2. This protein belongs to the d-isomer-specific 2-hydroxyacid dehydrogenase family and contains a NADH/NADPH-binding site. To provide biochemical evidence, we overexpressed the corresponding open reading frame in Escherichia coli and tested different substrate/cosubstrate combinations. The purified enzyme showed activity with both HP and Glx, and NADPH was the favored cosubstrate (Fig. 1). Moreover, oxalate inhibits the recombinant enzyme up to 70% for each substrate/cosubstrate combination. This inhibition was reported to be a characteristic for HPR2 but not for HPR1 (Kleczkowski et al., 1991; Timm et al., 2008). Concerning these findings, we conclude that the identified protein is not a typical HPR such as HPR1 but rather able to function similarly to HPR2; therefore, we termed it HPR3. Another candidate protein, At2g45630 (Timm et al., 2008), did not show activity with HP or Glx and is likely unrelated to photorespiration.
Figure 1.
Activity of recombinant At1g12550 (HPR3) and its inhibition by oxalate. A, Specific activity with HP and Glx using cofactors NADH and NADPH, respectively. B, Inhibition of the recombinant enzyme by oxalate (2 mm). Shown are mean activities ± sd from three independent measurements (μmol min−1 mg−1 protein). Asterisks show significant differences according to Student’s t test (P < 0.01).
Isolation of T-DNA Insertional Mutants of AtHPR3
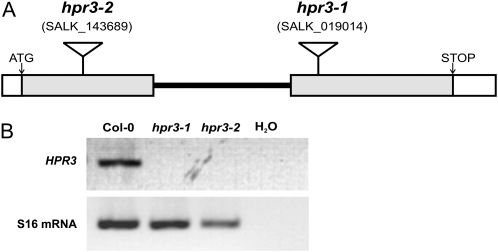
To examine the physiological function of HPR3, we set out to isolate two independent T-DNA insertion lines for the gene At1g12550. In these lines, hereafter designated as hpr3-1 and hpr3-2, insertions are located in the second exon and the first exon (Fig. 2A), respectively. First, homozygous lines were isolated by genomic PCR, and sequencing of the resulting fragments was obtained from both lines. The complete knockouts then were verified by reverse transcription (RT)-PCR analysis, using primer pairs that span the T-DNA insertion sites of the two loci. The 40S ribosomal protein S16 mRNA (At2g09990) was used as an internal control for the integrity of the generated cDNA. The mature transcript was found to be present in wild-type plants but was absent in both insertion mutants (Fig. 2B). These results confirmed the complete knockout of the At1g12550 gene in both T-DNA insertion lines.
Figure 2.
Isolation of HPR3 knockout mutants. A, Structure of the HPR3 gene and positions of the T-DNA insertions of two independent lines (gray bars represent the exons). B, HPR3-specific transcripts were undetectable in both homozygous lines but were found to be present in wild-type plants. Whole-leaf RNA was isolated and analyzed by RT-PCR using S16 mRNA as a control for the integrity of generated cDNA. This result was obtained for six plants (each line) and three technical replicates.
Knockout of AtHPR3 Does Not Impair Growth But Leads to Slightly Increased Levels of Photorespiratory Intermediates
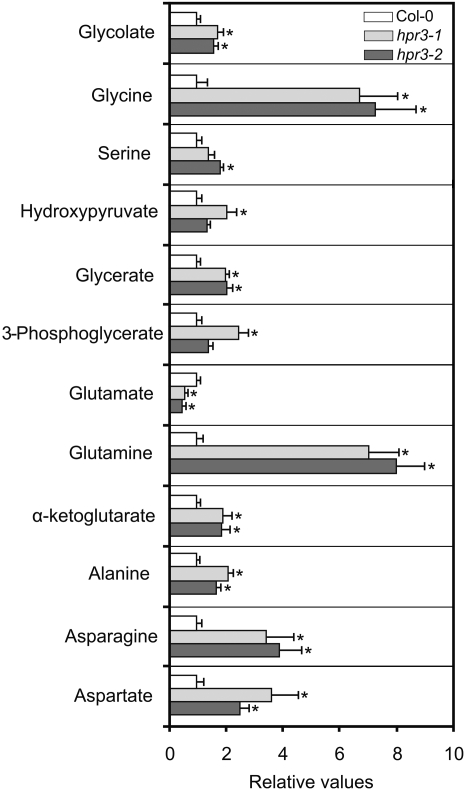
These HPR3-deficient mutant lines were grown on soil under different photoregimes to allow a comprehensive phenotypic analysis. However, no phenotypic differences were observed compared with the wild-type control, regardless of which photoperiod was used (10/14-h, 12/12-h, or 16/8-h day/night cycle). To examine the effect of knocking out HPR3 at the biochemical level, we next carried out metabolic analyses of the respective mutants. For this purpose, plants were grown on soil with a standard photoperiod of 10 h. After reaching growth stadium 5.1 (Boyes et al., 2001), samples were taken in the middle of the light period and analyzed by gas chromatography-mass spectrometry according to Lisec et al. (2006). Despite the wide range of unaltered metabolites (Supplemental Table S1), we found some slightly increased intermediates of the photorespiratory cycle, namely HP, 3PGA, glycerate, glycolate, and Gly (Fig. 3). We also found that these changes were accompanied by correspondingly altered fluxes, as assessed by the redistribution of label following feeding of leaf discs with [U-13C]Glc (Supplemental Table S2; Sienkiewicz-Porzucek et al., 2008). These alterations provide evidence that the photorespiratory flux is slightly disrupted and thus that the function of HPR3 is associated with photorespiration.
Figure 3.
Alteration of the leaf content of selected metabolites in the individual hpr3-1 and hpr3-2 knockout plants compared with the wild type (ecotype Columbia [Col-0]). Plants were grown in normal air with a 10/14-h day/night cycle. Leaves of six individual plants per line were harvested in the middle of the photoperiod at developmental stage 5.1 (Boyes et al., 2001). Mutant-to-wild-type ratios ± sd of relative mean metabolite contents are shown with the mean wild-type values arbitrarily set to 1 (wild type, white bars; hpr3-1, light gray bars; hpr3-2, dark gray bars). Asterisks show significant changes according to Student’s t test (P < 0.05).
Generation of a Triple HPR Mutant Leads to Increased Air Sensitivity But Fully Recovers in High-CO2 Conditions
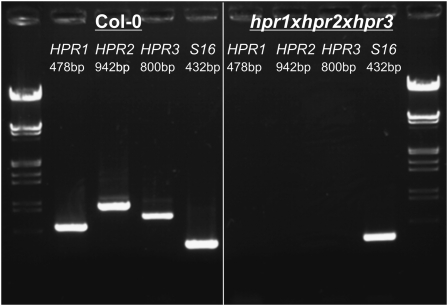
Given that the double knockout of HPR1 and HPR2 is still viable in air, multiple mutants were generated by integrating the newly identified HPR3 mutant by conventional crossing. For this purpose, the isolated line hpr3-1 was crossed with the previously isolated hpr1xhpr2 double knockout (Timm et al., 2008) to generate an HPR triple mutant and additionally to gain all other possible combinations of HPR double mutants. After crossing, the resulting T1 plants were characterized by genomic PCR. Thereafter, seeds were produced from the triple heterozygous intermediate, and plants again were grown under elevated-CO2 conditions (1% CO2 in air). This procedure was chosen to ensure that all homozygous combinations could be isolated, considering the conditional lethal phenotype of most other photorespiratory mutants (Boldt et al., 2005; Voll et al., 2006; Schwarte and Bauwe, 2007). Finally, a triple homozygous mutant of HPR1, HPR2, and HPR3 could by isolated under these conditions. The transcription of all three HPRs was analyzed by RT-PCR in order to verify the complete knockout of all HPRs. In the corresponding triple mutant, no transcripts were detectable from all three different genes, using S16 mRNA as an internal control for the integrity of the generated cDNA (Fig. 4).
Figure 4.
Crossing of hpr1-1, hpr2-1, and hpr3-1 leads to functional inactivation of the three HPR genes. RT-PCR analysis shows the absence of the transcripts encoded by the three HPR genes in leaves of the isolated triple mutant, whereas all three transcripts are present in the wild-type control. S16 mRNA was used as an internal control for the integrity of the produced cDNA. This result was obtained with three independent plants per line and two replicate RT-PCR analyses each.
As described above, during the crossing process of hpr1-1, hpr2-1, and hpr3-1, all possible combinations of double mutants also could be isolated. As visible in Figure 5B, the double mutant hpr1xhpr3 does not show any further growth retardation relative to the corresponding hpr1-1 single mutant, nor do the hpr2-1 and hpr3-1 single mutants and the hpr2xhpr3 double mutant show any retardation relative to the corresponding wild type. Only the hpr1xhpr2 double mutant shows a clear growth reduction compared with the hpr1-1 single mutant. As we can learn from the double knockouts including HPR3 (hpr1xhpr3 and hpr2xhpr3), this enzyme is only responsible for a small proportion of the HP-into-glycerate interconversion if HPR1 and HPR2 are present. However, the absence of all three HP-reducing enzymes further leads to strong impairment of growth under ambient air conditions compared with the double knockout of HPR1 and HPR2. Increased levels of CO2 in air (more than 0.15%) fully recover this oxygen-sensitive phenotype (Fig. 5C), which clearly demonstrates that the phenotype of the HPR triple mutant is related to photorespiration.
Figure 5.
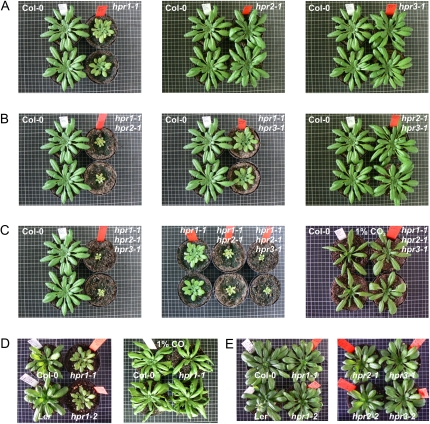
The combined deletion of HPR1, HPR2, and HPR3 leads to a significant decrease in plant growth but is recovered under high-carbon conditions. Plants were grown on soil in ambient air (except for C and D, right panel, which used 1% CO2) with a day/night cycle of 10/14 h (except for E, which used 12/12 h). Photographs were taken after 8 weeks of cultivation. A, Phenotypes of the HPR single mutants (from left to right: hpr1-1, hpr2-1, and hpr3-1) compared with the wild-type control (Col-0). B, Phenotypes of the generated HPR double mutants (from left to right: hpr1-1xhpr2-1, hpr1-1xhpr3-1, and hpr2-1xhpr3-1). C, Phenotype of the isolated HPR triple mutant in normal air (left panel) compared with HPR1 and the HPR1/HPR2 double mutant (middle panel) and under high-carbon conditions (1% CO2) compared with the wild-type control (right panel). D, A stronger phenotype is visible under short days (10/14 h; left panel) but could be compensated by cultivation in elevated CO2 (1%; right panel). E, This effect vanishes by cultivation with longer photoperiods (more than 12 h of light). HPR2 and HPR3 single mutants do not display this effect.
HPR Mutants Show Impaired Growth and Contain Less Chlorophyll
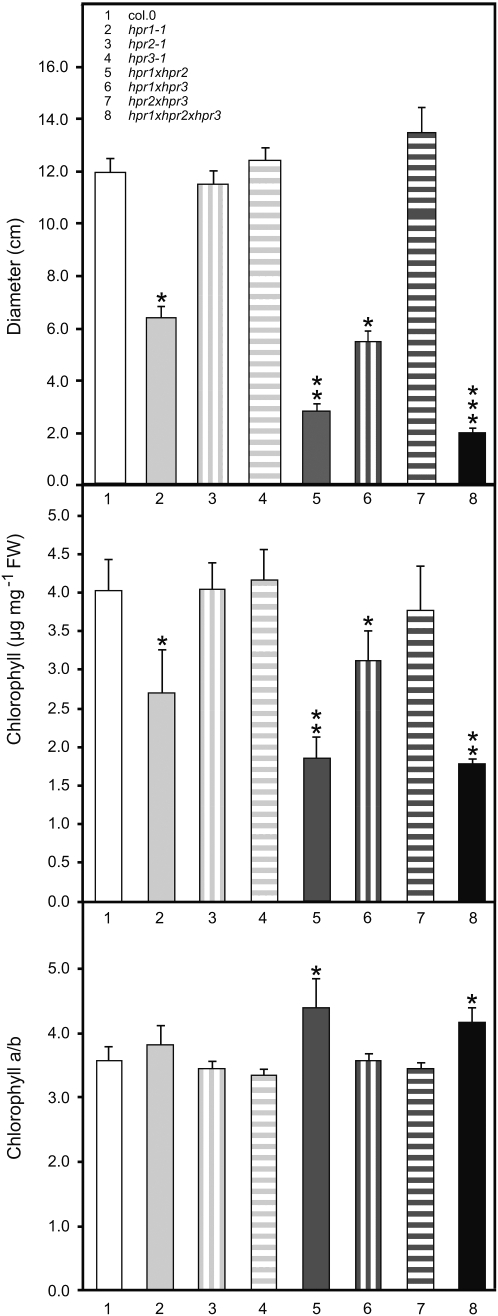
To further substantiate the visible impairment of growth, rosette diameters and chlorophyll contents were determined from short-day-grown plants (10/14-h day/night cycle). As visible in Figure 5 (A–C) and Figure 6, growth reduction is exacerbated by the increasing extent of HPR mutation. HPR1 mutants show growth inhibition up to 46% compared with the corresponding wild type under short-day conditions. This effect is not present with photoperiods longer than 12 h (Fig. 5E; Timm et al., 2008). The hpr1xhpr2 double knockout is inhibited up to 76% compared with the wild-type control and furthermore is reduced up to 57% compared with the hpr1 single mutant. The hpr1xhpr2xhpr3 triple mutant generated here is even more affected than the double mutant, with inhibition levels up to 83% in comparison with the wild-type control and a further growth retardation of up to 29% compared with the double mutant. In clear contrast to the growth reduction of the above-mentioned mutants, the single mutants hpr2 and hpr3 (compared with the wild type) as well as the double knockouts hpr1xhpr3 and hpr2xhpr3 (compared with hpr1 or hpr2 single mutant) displayed no growth effects.
Figure 6.
HPR multiple mutants show reduced growth and total chlorophyll content in ambient air. Plants were grown in ambient air with a 10/14-h day/night cycle. Measurements were performed after plants reached growth stadium 5.1 according to Boyes et al. (2001). Rosette diameters were measured as the longest distance (in cm) across fully expanded leaves. Mean values ± sd are shown from five independent determinations each. Asterisks show significant changes according to Student’s t test either in comparison with the corresponding wild type (* P < 0.05), with the single mutants (** P < 0.05), or with the hpr1xhpr2 double mutant (*** P < 0.05). FW, Fresh weight.
Closely corresponding to the growth impairment, total chlorophyll contents are reduced in HPR mutants (Fig. 6). The hpr1 knockout has 35% less total leaf chlorophyll content, while the hpr1xhpr2 double mutant contains 49% less total chlorophyll than the wild type. This value is also decreased yet further in the hpr1xhpr2xhpr3 triple mutant but not significantly so in comparison with the hpr1xhpr2 double mutant. Furthermore, the leaf chlorophyll a/b ratio is also increased in the hpr1xhpr2 double mutant and in the triple mutant, indicating a senescence-mediated chlorophyll breakdown (Pruzinská et al., 2005) that is related to the elevated oxygen sensitivity. However, neither hpr2 and hpr3 single mutant nor the corresponding double mutant hpr2xhpr3 are affected in their chlorophyll contents or chlorophyll a/b ratios. The third double mutant, hpr1xhpr3, only shows reduced levels in chlorophyll content comparable to hpr1 single mutants. These results once more clearly reflect that HPR1 and HPR2 are the most active isoforms in Arabidopsis.
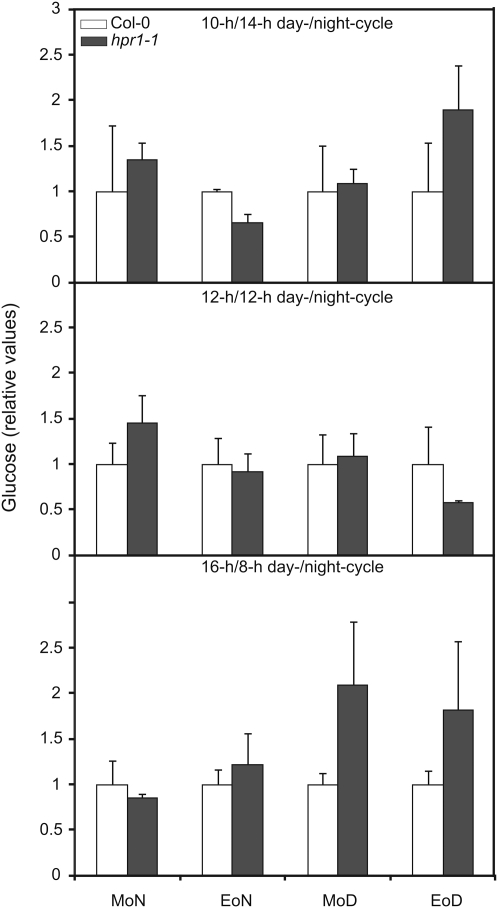
As illustrated in Figure 5 (A and D, left panel), hpr1 single mutants also displayed a growth inhibition phenotype under conditions in which the photoperiod is shorter than 12 h of light. This effect is an exclusive characteristic of hpr1 mutants, with hpr2 and hpr3 mutants not displaying such a photoperiodic effect on growth (Fig. 5E, right panel). By contrast, the effect in hpr1 single mutants vanishes upon light periods longer than 12 h of light (Fig. 5E, left panel; Timm et al., 2008). This result possibly could be explained by carbon starvation of the Calvin cycle under shorter photoperiods during the prolonged night (Matt et al., 1998). The photorespiratory flux is clearly not optimal in hpr1 mutants under these conditions, which enhances the general short-day effect described above and as suggested by decreased Glc levels at the end of the night under short-day conditions (Fig. 7). Moreover, this effect could be reversed by cultivation in high-carbon conditions (greater than 0.15% CO2), clearly indicating that a disrupted photorespiratory cycle is responsible for this daylength-dependent phenotype (Fig. 5D, right panel).
Figure 7.
HPR1 mutants show a reduced Glc level at the end of the night under shorter photoperiods. Plants were grown in normal air with 10/14-h, 12/12-h, and 16/8-h day/night cycles. Leaves of three individual plants at developmental stage 5.1 (Boyes et al., 2001) were harvested in the middle (MoD) and the end (EoD) of the day and the middle (MoN) and the end (EoN) of the night. Mutant-to-wild-type ratios of mean relative metabolite contents ± sd are shown (n = 3), with the mean wild-type values arbitrarily set to 1.
Photochemical Efficiencies Differ According to the Number of Deleted HPRs
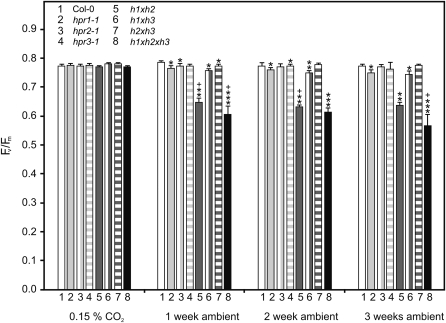
It is well known that repair mechanisms of PSII are inhibited as a result of an interruption of the Calvin cycle (Long et al., 1994; Takahashi and Murata, 2005). To further substantiate the effects of HPR mutations on PSII activity and hence limitations to the Calvin cycle, we measured the photochemical efficiency (maximum variable fluorescence/maximum yield of fluorescence [Fv/Fm]) of PSII after different periods of exposure of the mutants to ambient air (Fig. 8). Whereas wild-type plants did not show any response, slightly decreased values were observed for the hpr1 single mutant and distinctly stronger effects for the hpr1xhpr2 double knockout and the hpr1xhpr2xhpr3 triple mutant. This effect was exacerbated with the length of exposure to ambient air and is further exaggerated in the triple mutant. In contrast, neither the hpr2 and hpr3 single mutants nor the corresponding hpr2xhpr3 double knockout showed a response in this parameter. The double mutant hpr1xhpr3 displayed values comparable to hpr1 single mutants. Compared with other photorespiratory mutants, in which the Fv/Fm ratio responds very rapidly after exposure to ambient air (Takahashi et al., 2007b), the respective HPR mutants showed some degree of long-term response to air levels of oxygen. This might be due to the relatively moderate metabolic alterations when compared with the changes observed after knockout of other photorespiratory enzymes.
Figure 8.
HPR multiple mutants show decreased Fv/Fm values after transfer to ambient air. Plants were grown in elevated CO2 (0.15%) with a 10/14-h day/night cycle. After reaching growth stadium 5.1 (Boyes et al., 2001), plants were transferred to ambient air. Fv/Fm ratios were measured before transfer and 1, 2, and 3 weeks after transfer to normal air. Mean values ± sd (n = 5; five areas of interest each) are shown. Asterisks show significant alterations according to Student’s t test (P < 0.05 for the wild type [*], hpr1-1 [**], hpr1xhpr2 [***], and the corresponding line 1 week before transfer [+]).
Photosynthetic Parameters Differ According to the Number of Deleted HPRs
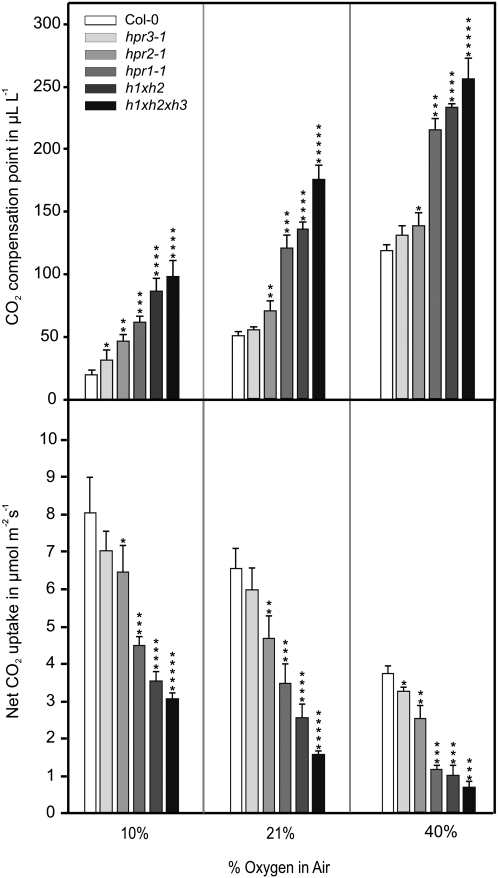
To further characterize the response of HPR mutants to various oxygen levels, which directly affect photorespiratory flux, we determined photosynthetic rates and CO2 compensation points (γ) at three different oxygen concentrations (Fig. 9; Supplemental Table S3). The second parameter, γ, depends on the balance between photosynthetic CO2 uptake and respiratory CO2 release, and in C3 plants, its values respond linearly to changing oxygen concentrations (Farquhar et al., 1980). Since most of the multiple knockouts show a distinct growth inhibition in air and an early onset of flowering, HPR mutants were first cultivated under mild nonphotorespiratory conditions (0.15% CO2) to synchronize growth and then adapted to ambient air for 1 week. Relative to wild-type plants, we observed distinct though not always significant changes to both parameters of photosynthetic gas exchange in the order hpr3 < hpr2 < hpr1. This finding indicates inferior but yet recognizable roles of HPR2 and HPR3 for photorespiratory metabolism as long as HPR1 is active. In the absence of HPR1, the additional deletion of HPR2 results in a massive negative impact on photosynthesis that is even more pronounced in the hpr1xhpr2xhpr3 triple mutant. While such effects were observed at all three oxygen concentrations, they were most noticeable at the highest oxygen concentration. Additionally, the calculation of the slope (γ) from the estimatedГ/oxygen response lines also indicates an ascending negative effect on photosynthesis, according to the number of deleted HPRs. The value of γ increased in the order wild type < hpr1 < hpr1xhpr2 < hpr1xhpr2xhpr3 (numbers are given in the legend to Fig. 9) if plants were first grown in high-carbon conditions and then acclimatized to air conditions for 1 week. Control experiments confirmed that neither HP nor Ser increased, nor were γ values altered in the mutants relative to the wild type before the transfer to normal air (data not shown).
Figure 9.
Oxygen-dependent gas exchange of selected HPR mutants. Plants were grown in elevated CO2 (0.15%) with a 10/14-h day/night cycle. After reaching growth stadium 5.1 (Boyes et al., 2001), plants were transferred to ambient air and, after 1 week of adaptation, photosynthetic rates and CO2 compensation points were measured as a function of oxygen concentration (10%, 21%, and 40%). Mean values ± sd (n = 5) are shown and are listed in Supplemental Table S3. Asterisks show significant alterations according to Student’s t test (P < 0.05 for the wild type [*], hpr3-1 [**], hpr2-1 [***], hpr1-1 [****], and hpr1xhpr2 [*****]). Calculated slopes of the γ/oxygen response lines were 0.000355 ± 0.000032 μL μL−1 (wild type), 0.000507 ± 0.000028 μL μL−1 (hpr1), 0.000531 ± 0.000024 μL μL−1 (hpr1xhpr2), and 0.000561 ± 0.000018 μL μL−1 (hpr1xhpr2xhpr3).
Leaves Accumulate Ser and Related Metabolites
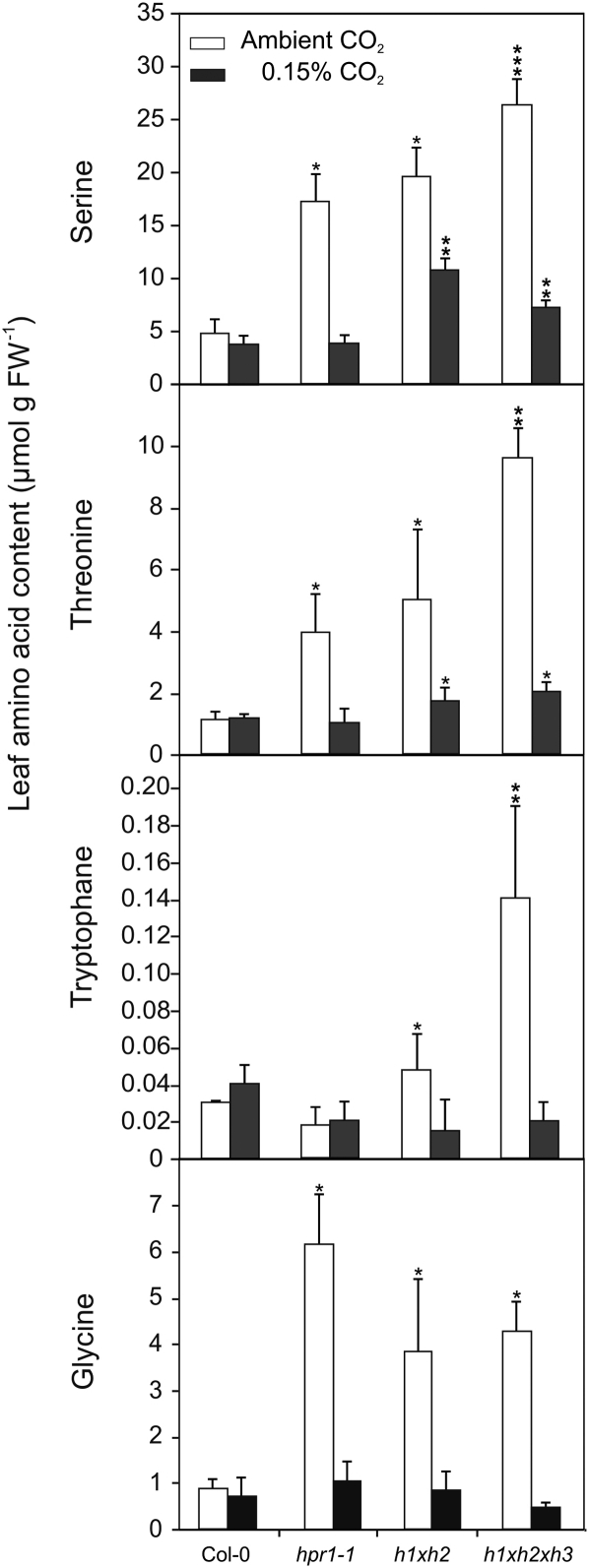
To substantiate knockout-related effects under photorespiratory and nonphotorespiratory conditions, we examined the levels of leaf amino acids using plants grown in ambient air and elevated CO2 (0.15%). The most striking observation was the strong accumulation of the photorespiratory intermediate Ser (Fig. 10). The effect is consistent and rises with the level of HPR inhibition, reaching as high as 26 μmol g−1 fresh weight in the hpr1xhpr2xhpr3 triple mutant. However, not only Ser but also the metabolically related amino acids Thr and Trp accumulate, suggesting an up-regulation of Ser-metabolizing processes. Additionally, Gly, the direct precursor of Ser within the photorespiratory cycle, also accumulates, underlining altered Ser metabolism; however, this effect is not increased with the level of HPR inhibition. Given that nearly all of the described metabolic effects could be reversed by elevated CO2, it seems reasonable to assume that they result from the disrupted photorespiratory flux.
Figure 10.
Leaf amino acid analysis reveals strong accumulation of Ser and related metabolites. Plants were grown in elevated CO2 (0.15%) and in normal air with a 10/14-h day/night cycle. Leaves from six individual plants at developmental stage 5.1 (Boyes et al., 2001) were harvested in the middle of the photoperiod, and soluble amino acids were analyzed by HPLC. Mean amino acid contents ± sd are shown. Asterisks indicate significant changes according to Student’s t test (P < 0.05 for the corresponding wild-type sample [*], hpr1-1 [**], and hpr1xhpr2 [***]). FW, Fresh weight.
DISCUSSION
During the last years, the core enzymes of the photorespiratory cycle have been placed under intensive analyses at both the molecular and biochemical levels (for review, see Bauwe et al., 2010). With the exception of the multigene-encoded glycolate oxidase, Arabidopsis mutants could be isolated for all of these enzymes. Taken together, such mutants share one common feature, their strong sensitivity to air, which can easily be reverted by cultivation in elevated concentrations of CO2. This situation is long known from studies with chemically generated mutants (Somerville, 2001), but, more recently, it was shown that several exceptions exist. First, high-CO2 conditions cannot rescue mutants without Gly decarboxylase activity (Engel et al., 2007). This is because this particular enzyme not only functions in the photorespiratory cycle but also in one-carbon metabolism and therefore is absolutely necessary for plant development under any conditions. Second, we previously showed that, contrary to what was commonly thought, photorespiratory HP reduction is not restricted to peroxisomes but, via HPR2, can also occur in the cytosol, resulting in an atypically low air sensitivity of hpr1 mutants (Timm et al., 2008). Third, photorespiration has specific links to folate (Collakova et al., 2008) and energy metabolism (Sweetlove et al., 2006). The photorespiratory pathway is thus integrated into whole-cell metabolism in a more complex manner than previously considered. While most of the other photorespiratory mutants are not very useful to investigate connections with other pathways, on account of their conditional lethal phenotypes in ambient air, the HPR mutants are not constrained in this manner. Utilizing the moderate phenotype of HPR mutants, connections to other pathways could be studied and compared with the wild-type situation. Using this approach, we observed that knocking out HPR1 and HPR2 invokes effects that are not restricted to photorespiration. In this previous study, we observed links to tricarboxylic acid cycle intermediates, amino acid metabolism, and ethanolamine, which acts as a precursor of choline biosynthesis (Timm et al., 2008). Such cross talk and connections are still under investigation, but the HPR mutants appear to be a very useful tool to address such interactions.
This study presents evidence that HP reduction is not even restricted to peroxisomes and cytosol but involves chloroplasts too, albeit to a lesser extent. By the identification of a novel 2-hydroxyacid dehydrogenase as an enzyme with HP and Glx reductase activity (Fig. 1), we could enlarge the general importance of the reduction of these two metabolites. Since the newly identified HPR3 is predicted to be located in chloroplasts (http://aramemnon.botanik.uni-koeln.de/tm_sub.ep?GeneID=13660&modelID=0; Schwacke et al., 2003; Yu et al., 2008), a third compartment is likely involved in the reduction of these photorespiratory intermediates. As we show by metabolite analyses of HPR3 single knockouts (Fig. 3), some photorespiratory intermediates (glycolate, Gly, Ser, 3HP, and glycerate) are slightly altered, which indicates a disruption of the photorespiratory cycle, thus further cementing the association of HPR3 function to that of photorespiration. Furthermore, we discovered changes in the levels of α-ketoglutarate, Glu, and Gln, which indicate a shift in the carbon-nitrogen balance of the mutants. These effects are consistent with those observed following the knockout of other photorespiratory genes (Igarashi et al., 2006; Eisenhut et al., 2008; Timm et al., 2008). The higher levels of Ala, Asn, and Asp additionally indicate an impairment in nitrogen assimilation and may be regulated by the increased levels of Gln. The altered Thr level could further support this notion or alternatively could be due to the conversion of the Ser, which accumulates, to alternative metabolites. Moreover, the combined deletion of HPR1, HPR2, and HPR3 leads to strong air sensitivity, which, as mentioned above, is a cardinal property of all other photorespiratory mutants. In this triple mutant, not only the CO2 compensation point and net CO2 uptake were strongly inversely influenced (Fig. 9) but also the photochemical efficiency of PSII after transfer of the mutant from elevated to air levels of CO2 (Fig. 8).
This latter observation corresponds well to a previous report that impairment of the photorespiratory cycle and, as a consequence, the Calvin cycle leads to pronounced photoinhibition by suppression of the synthesis of the D1 subunit of PSII (Takahashi et al., 2007a). Apart from this impact on linear electron transport, elevated levels of photorespiratory metabolites can inhibit carbon flux through the Calvin cycle. This is not restricted to the 2PG effects mentioned in the introduction. It is also known, for example, that Glx inhibits Rubisco activase and hence Rubisco (Chastain and Ogren, 1989; Campbell and Ogren, 1990; Häusler et al., 1996), and Gly accumulation is toxic due to binding of Mg2+ by complex formation (Eisenhut et al., 2007). In addition, the elevated oxygen dependency of γ in all HPR mutants indicates higher rates of decarboxylation of photorespiratory metabolites, for example Ser (Rontein et al., 2003) or HP (Hedrick and Sallach, 1961), outside the core photorespiratory cycle. This is because the slope γ of the γ-versus-oxygen response curve is essentially codetermined by two factors, the Rubisco specificity factor, which is invariable in a given species, and the stoichiometry (i.e. tightness) of the photorespiratory cycle (Farquhar et al., 1980). This stoichiometry of ribulose 1,5-bisphosphate oxygenation versus photorespiratory CO2 release, which would be exactly 2 if only Gly becomes decarboxylated, apparently shows some variation dependent on the fraction of additional decarboxylation reactions. This fraction could well be enhanced by environmental conditions such as higher light intensities and/or temperatures (Hanson and Peterson, 1986) and the intactness of photorespiratory metabolism (this work). Interestingly, mutants of the peroxisomal malate dehydrogenases support these results, because oxygen-dependent γ values are also changed in these plants (Cousins et al., 2008). Concerning the fact that both isoforms are responsible for NADH supply of the peroxisomes, the double knockout is quite similar to HPR1 mutants and seems to be a good confirmation of the results presented here. Unfortunately, no other defined photorespiratory mutants of Arabidopsis have been analyzed in this context, and it remains open if this is an exclusive feature of mutants with a defective HP-to-glycerate conversion step.
The above-mentioned conclusion is further substantiated by the analysis of the leaf amino acid content of the mutants, which revealed a distinct accumulation of the photorespiratory intermediate Ser (Fig. 10). This is perhaps not overly surprising, since this amino acid is the direct precursor of HP (Liepman and Olsen, 2001), but the enhanced levels could indeed trigger additional decarboxylating reactions, most prominently to ethanolamine, as we have shown previously (Timm et al., 2008), and therefore affect γ in HPR mutants. Nevertheless, this enhancement is not restricted only to Ser but also includes the metabolically related amino acids Thr and Trp and additionally influences the steady-state level of Gly. The latter is consistently increased in all mutants but showed no response to the level of HPR mutation.
In addition to increased decarboxylating reactions of photorespiratory metabolites, the ratio of respiration in the light to the maximum rate of Rubisco carboxylation could also influence γ. This also seems to be a likely explanation for the altered values in HPR mutants. This notion is supported by a previously radiogasometric study of HPR1 mutants, which show higher respiratory rates of primary and stored photosynthates in the dark and the light (supplemental table S1 in Timm et al., 2008; Pärnik and Keerberg, 2007). It seems reasonable that these reactions become pronounced with the level of HPR mutation and therefore lead to increased values of γ.
As we previously demonstrated, knocking out HPR1 does not lead to the typical phenotype associated with the knockout of most other photorespiratory enzymes (Timm et al., 2008). Interestingly, given the daylength-dependent phenotype we demonstrated here (Figs. 5 and 7), it seems likely that gene redundancy is only part of the reason behind this phenomenon. It is a general observation that plants that are grown in short days become carbon starved during a prolonged night (Matt et al., 1998; Graf et al., 2010). This effect is exacerbated in HPR1 mutants, also indicating that there is a possible connection of photorespiration and carbon fixation during the Calvin cycle. Most likely, this is mediated by reduced 3PGA recycling resulting from the disrupted photorespiratory flux in combination with an outflow of metabolites from the cycle. The photorespiratory flux is clearly not optimal in hpr1 mutants under these conditions, which enhances the general short-day effect described above and as suggested by decreased Glc levels at the end of the night under short-day conditions (Fig. 7).
Here, we report to our knowledge a previously unknown enzyme that is able to reduce both HP and Glx, two of the key metabolites within the photorespiratory cycle, and hence could directly or indirectly contribute to photorespiration in higher plants. Considering its dual substrate specificity and its preference for NADPH as the cosubstrate, the identified HPR3 could support peroxisomal and cytosolic HPRs (Timm et al., 2008) as well as chloroplastidial and cytosolic glyoxylate reductase (Allan et al., 2009) for an optimal reduction of these intermediates. Given the reported chloroplastic localization of HPR3 (Yu et al., 2008), its exact physiological role is not absolutely clear; however, the observed effects on photosynthesis (Fig. 9) and the normalization of the elevated levels of several diagnostic amino acids by elevated CO2 (Fig. 10) clearly suggest that its function is indeed associated with photorespiration. Unfortunately, no HP transporter was identified yet (Reumann and Weber, 2006); therefore, it is not known how this photorespiratory intermediate gets into chloroplast. Since the chemical structure of HP is similar to glycerate, it can be hypothesized that import could possibly occur via the reported glycerate/glycolate transporter (Howitz and McCarty, 1986). In addition to the reduction of HP, the newly identified HPR3 could also contribute to the reduction of chloroplastidially generated Glx, which, as discussed above, is an inhibitor of several enzymes involved in photosynthetic CO2 fixation. This, however, is not very likely, since a plastidial Glx reductase exists (Simpson et al., 2008) and the single knockout of HPR3 does not cause drastic effects (Fig. 5).
The construction of a chloroplast protein interaction network in Arabidopsis emphasizes an additional role of HPR3 in plant metabolism (Yu et al., 2008). From these analyses, it appears that HPR3 could interact with three other chloroplast proteins, two of which, 3PGA dehydrogenase (At1g17745) and phosphoserine aminotransferase (At4g35630), are related to Ser synthesis. The third interacting protein, Ser hydroxymethyltransferase (At4g32520), is related to Gly synthesis and one-carbon metabolism (Zhang et al., 2010). Thus, HP could be generated in glycolytic routes from 3PGA, most likely the phosphoserine pathway. This pathway is best described in bacteria but also is found in plants, mainly in heterotrophic issue or during the night when the photorespiratory flux is not active (Ho et al., 1998; Muñoz-Bertomeu et al., 2009). Supportive evidence for this hypothesis is the accumulation of Gly, which we observed in the HPR3 single mutants. If HPR3 works in the phosphoserine pathway, the deletion of this enzyme would be expected to lead to a disrupted flow, which would explain the altered Gly-to-Ser interconversion in the mutants. However, the exact mechanism behind these changes is not yet known, and it would be very informative to analyze mutants within the phosphoserine pathway under nonphotorespiratory and photorespiratory conditions as well as in combination with different HPR mutations. It seems likely that alternative Ser-synthesizing pathways, like the phosphoserine-metabolizing route, interact with the photorespiratory pathway and, therefore, the HPR triple mutant shows further growth retardation.
In conclusion, although the role for HPR3 will need further investigation, this study presents strong indications that the enzyme is associated with the photorespiratory process and can at least partially complement the function of the peroxisomal HPR1 and the cytosolic HPR2. Moreover, HPR3 could display a possible link of photorespiration to other Ser-synthesizing pathways.
MATERIALS AND METHODS
Plant Material and Growth
Arabidopsis (Arabidopsis thaliana) ecotypes Columbia and Landsberg erecta were used as wild-type references. SALK lines SALK_067724 (hpr1-1), SALK_143584 (hpr1-2), SALK_105875 (hpr2-1), SALK_019014 (hpr3-1), and SALK_143689 (hpr3-2) and gene trap line GT.89166 (hpr2-2) were obtained from the Nottingham Arabidopsis Stock Centre (Sundaresan et al., 1995; Alonso et al., 2003). Seeds were incubated at 4°C for at least 2 d to break dormancy prior to germination. Plants were grown on soil (Type Mini Tray; Einheitserdewerk) and vermiculite (4:1 mixture) and were regularly watered with 0.2% Wuxal liquid fertilizer (Aglukon). Unless otherwise stated, plants were grown in normal air (380–400 μL L−1 CO2) and in air with elevated CO2 (0.15% or 1% as indicated in the figure legends) in controlled-environment chambers (Percival) with a day/night cycle of 10/14 h (22°C/18°C, approximately 120 μmol m−2 s−1 irradiance).
Knockout Mutants and RT-PCR
Insertion lines for HPR1 (hpr1-1 and hpr1-2) and HPR2 (hpr2-1 and hpr2-2) were described previously (Timm et al., 2008). For T-DNA insertion lines of HPR3, leaf DNA was isolated and PCR amplified (1 min at 94°C, 1 min at 58°C, 1 min at 72°C; 35 cycles) with specific left border primers (R295 for hpr3-1 and R175 for hpr3-2) and a corresponding primer for the open reading frame of At1g12550 (R669) to verify insertions. The nucleotide sequences of all primers used in this study are shown in Supplemental Table S4. Zygosity was then validated using leaf DNA and specific primers for the genomic region of At1g12550 (R665 and R669). The knockout of HPR3 was verified by RT-PCR using 2.5 μg of leaf RNA for cDNA synthesis (Nucleospin RNA plant kit [Macherey-Nagel] and RevertAid cDNA synthesis kit [MBI Fermentas]) and the primer combination P424/P425, yielding a 972-bp fragment. This fragment was present in the wild type but not in homozygous knockout lines. The amount of cDNA for PCR was calibrated according to signals from 432-bp fragments obtained by PCR amplification of the constitutively expressed 40S ribosomal protein S16 gene, with primers R176 (sense) and R177 (antisense) before PCR analysis.
Multiple HPR mutants were generated by crossing the HPR1 and HPR2 double knockout (Timm et al., 2008) with the isolated line hpr3-1, yielding a triple heterozygous intermediate line for HPR1, HPR2, and HPR3. This line then was grown under elevated CO2 concentrations (1%). Leaf DNA was isolated and analyzed by genomic PCR to verify the presence of all three insertions and zygosity using the primers listed in Timm et al. (2008) for HPR1 and HPR2 and in Supplemental Table S4 for HPR3. The complete knockout of HPRs was double checked by RT-PCR as described above, yielding two double mutants, hpr1-1xhpr3-1 and hpr2-1xhpr3-1, and the corresponding triple mutant hpr1-1xhpr2-1xhpr3-1.
Heterologous Expression
The entire coding region of At1g12550 transcript was amplified from leaf RNA by RT-PCR with primers R987 and R988 (Supplemental Table S4) under simultaneous introduction of two flanking SacII restriction sites. The PCR product was first cloned into pGEM T-Easy (Promega) and then, after sequence confirmation and using the flanking SacII restriction sites, inserted in-frame into the coding sequence of the N-terminal His tag of the Escherichia coli overexpression vector pHUE (Catanzariti et al., 2004). Recombinant HPR3 was purified using nickel-nitrilotriacetic acid agarose (HisTrap) and Q-Sepharose blue affinity chromatography (GE Healthcare).
Chlorophyll Determination and PSII Fluorescence
Leaf chlorophyll content was determined as described by Porra (2002) and Porra et al. (1989). The Fv/Fm ratio corresponding to the potential quantum yield of photochemical reactions of PSII was measured (Schreiber et al., 1986) using an imaging pulse amplitude modulation fluorometer (M series; Walz). Plants were first grown for 8 weeks under elevated CO2 conditions (1%) and then transferred to ambient air (10/14-h day/night cycle). Measurements were performed before (high-carbon control) and after transfer to ambient air (after 1, 2, and 3 weeks) as described by Fahnenstich et al. (2008).
CO2 Exchange
Gas exchange (net photosynthetic rate and CO2 compensation point) was analyzed with a Li-Cor-6400 gas exchange system (LI-COR) using fully developed rosette leaves from plants grown in normal air or air with elevated CO2 (0.15%) to developmental stage 5.1 as described above. Measurements were performed at a photosynthetic photon flux density of 250 μmol m−2 s−1, supplied by an in-built red/blue light-emitting diode light source, 380 μL L−1 CO2, 21% O2, and a leaf temperature of 25°C. Alternative oxygen concentrations (10% and 40%) were generated using the gas-mixing system GMS600 (QCAL Messtechnik). Plants were adapted to the respective conditions for at least 20 min before the actual measurement.
Metabolomics and Leaf Soluble Amino Acids
Metabolite analysis was performed using rosette leaves from plants at developmental stage 5.1 according to Boyes et al. (2001). Extraction and analysis were performed as described by Lisec et al. (2006). For amino acid determination, 100 mg of leaf material was ground in liquid nitrogen and extracted in 1.8 mL of 80% ethanol for 30 min. After centrifugation, the supernatant was vacuum dried and dissolved in 8 mm sodium phosphate (pH 6.8) and 0.4% tetrahydrofurane. Individual amino acids were separated by HPLC and quantified as described earlier (Hagemann et al., 2005).
Sequence data from this article can be found in the Arabidopsis Genome Initiative (2000) or GenBank/EMBL data libraries under the following accession numbers: At1g68010 (HPR1), At1g79870 (HPR2), At1g12550 (HPR3), and At2g09990 (40S ribosomal protein S16).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table S1. Relative levels of selected metabolites in the single mutants of HPR3.
Supplemental Table S2. Redistribution of label following feeding of leaf discs with [U-13C]Glc in the individual hpr3-1 and hpr3-2 knockout plants.
Supplemental Table S3. Oxygen-dependent gas-exchange parameters of HPR mutants.
Supplemental Table S4. Sequences of primers used in this study.
Supplementary Material
Acknowledgments
We are especially grateful to Martin Hagemann (University of Rostock) for helpful comments on the research. The excellent technical assistance with HPLC analysis from Klaudia Michl (University of Rostock) is acknowledged. We further thank the Nottingham Arabidopsis Stock Centre for providing us with the SALK lines to make this work possible.
References
- Allan WL, Clark SM, Hoover GJ, Shelp BJ. (2009) Role of plant glyoxylate reductases during stress: a hypothesis. Biochem J 423: 15–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Anderson LE. (1971) Chloroplast and cytoplasmic enzymes. II. Pea leaf triose phosphate isomerases. Biochim Biophys Acta 235: 237–244 [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Bauwe H, Hagemann M, Fernie AR. (2010) Photorespiration: players, partners and origin. Trends Plant Sci 15: 330–336 [DOI] [PubMed] [Google Scholar]
- Boldt R, Edner C, Kolukisaoglu Ü, Hagemann M, Weckwerth W, Wienkoop S, Morgenthal K, Bauwe H. (2005) d-GLYCERATE 3-KINASE, the last unknown enzyme in the photorespiratory cycle in Arabidopsis, belongs to a novel kinase family. Plant Cell 17: 2413–2420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyes DC, Zayed AM, Ascenzi R, McCaskill AJ, Hoffman NE, Davis KR, Görlach J. (2001) Growth stage-based phenotypic analysis of Arabidopsis: a model for high throughput functional genomics in plants. Plant Cell 13: 1499–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell WJ, Ogren WL. (1990) Glyoxylate inhibition of ribulosebisphosphate carboxylase/oxygenase activation in intact, lysed, and reconstituted chloroplasts. Photosynth Res 23: 257–268 [DOI] [PubMed] [Google Scholar]
- Catanzariti AM, Soboleva TA, Jans DA, Board PG, Baker RT. (2004) An efficient system for high-level expression and easy purification of authentic recombinant proteins. Protein Sci 13: 1331–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chastain CJ, Ogren WL. (1989) Glyoxylate inhibition of ribulosebisphosphate carboxylase/oxygenase activation state in vivo. Plant Cell Physiol 30: 937–944 [Google Scholar]
- Collakova E, Goyer A, Naponelli V, Krassovskaya I, Gregory JF, III, Hanson AD, Shachar-Hill Y. (2008) Arabidopsis 10-formyl tetrahydrofolate deformylases are essential for photorespiration. Plant Cell 20: 1818–1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cousins AB, Pracharoenwattana I, Zhou W, Smith SM, Badger MR. (2008) Peroxisomal malate dehydrogenase is not essential for photorespiration in Arabidopsis but its absence causes an increase in the stoichiometry of photorespiratory CO2 release. Plant Physiol 148: 786–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenhut M, Bauwe H, Hagemann M. (2007) Glycine accumulation is toxic for the cyanobacterium Synechocystis sp. strain PCC 6803, but can be compensated by supplementation with magnesium ions. FEMS Microbiol Lett 277: 232–237 [DOI] [PubMed] [Google Scholar]
- Eisenhut M, Huege J, Schwarz D, Bauwe H, Kopka J, Hagemann M. (2008) Metabolome phenotyping of inorganic carbon limitation in cells of the wild type and photorespiratory mutants of the cyanobacterium Synechocystis sp. strain PCC 6803. Plant Physiol 148: 2109–2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel N, van den Daele K, Kolukisaoglu Ü, Morgenthal K, Weckwerth W, Pärnik T, Keerberg O, Bauwe H. (2007) Deletion of glycine decarboxylase in Arabidopsis is lethal under nonphotorespiratory conditions. Plant Physiol 144: 1328–1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahnenstich H, Scarpeci TE, Valle EM, Flugge UI, Maurino VG. (2008) Generation of hydrogen peroxide in chloroplasts of Arabidopsis overexpressing glycolate oxidase as an inducible system to study oxidative stress. Plant Physiol 148: 719–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farquhar GD, von Caemmerer S, Berry JA. (1980) A biochemical model of photosynthetic CO2 assimilation in leaves of C3 species. Planta 149: 78–90 [DOI] [PubMed] [Google Scholar]
- Graf A, Schlereth A, Stitt M, Smith AM. (2010) Circadian control of carbohydrate availability for growth in Arabidopsis plants at night. Proc Natl Acad Sci USA 107: 9458–9463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemann M, Vinnemeier J, Oberpichler I, Boldt R, Bauwe H. (2005) The glycine decarboxylase complex is not essential for the cyanobacterium Synechocystis sp. strain PCC 6803. Plant Biol (Stuttg) 7: 15–22 [DOI] [PubMed] [Google Scholar]
- Hanson KR, Peterson RB. (1986) Regulation of photorespiration in leaves: evidence that the fraction of ribulose bisphosphate oxygenated is conserved and stoichiometry fluctuates. Arch Biochem Biophys 246: 332–346 [DOI] [PubMed] [Google Scholar]
- Häusler RE, Bailey KJ, Lea PJ, Leegood RC. (1996) Control of photosynthesis in barley mutants with reduced activities of glutamine synthetase and glutamate synthase. 3. Aspects of glyoxylate metabolism and effects of glyoxylate on the activation state of ribulose-1,5-bisphosphate carboxylase-oxygenase. Planta 200: 388–396 [Google Scholar]
- Hedrick JL, Sallach HJ. (1961) The metabolism of hydroxypyruvate. I. The nonenzymatic decarboxylation and autoxidation of hydroxypyruvate. J Biol Chem 236: 1867–1871 [PubMed] [Google Scholar]
- Ho CL, Noji M, Saito M, Yamazaki M, Saito K. (1998) Molecular characterization of plastidic phosphoserine aminotransferase in serine biosynthesis from Arabidopsis. Plant J 16: 443–452 [DOI] [PubMed] [Google Scholar]
- Howitz KT, McCarty RE. (1986) d-Glycerate transport by the pea chloroplast glycolate carrier: studies on [1-14C]D-glycerate uptake and D-glycerate dependent O2 evolution. Plant Physiol 80: 390–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi D, Miwa T, Seki M, Kobayashi M, Kato T, Tabata S, Shinozaki K, Ohsumi C. (2003) Identification of photorespiratory glutamate:glyoxylate aminotransferase (GGAT) gene in Arabidopsis. Plant J 33: 975–987 [DOI] [PubMed] [Google Scholar]
- Igarashi D, Tsuchida H, Miyao M, Ohsumi C. (2006) Glutamate:glyoxylate aminotransferase modulates amino acid content during photorespiration. Plant Physiol 142: 901–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly GJ, Latzko E. (1976) Inhibition of spinach-leaf phosphofructokinase by 2-phosphoglycollate. FEBS Lett 68: 55–58 [DOI] [PubMed] [Google Scholar]
- Kleczkowski LA, Randall DD, Edwards GE. (1991) Oxalate as a potent and selective inhibitor of spinach (Spinacia oleracea) leaf NADPH-dependent hydroxypyruvate reductase. Biochem J 276: 125–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liepman AH, Olsen LJ. (2001) Peroxisomal alanine:glyoxylate aminotransferase (AGT1) is a photorespiratory enzyme with multiple substrates in Arabidopsis thaliana. Plant J 25: 487–498 [DOI] [PubMed] [Google Scholar]
- Lisec J, Schauer N, Kopka J, Willmitzer L, Fernie AR. (2006) Gas chromatography mass spectrometry-based metabolite profiling in plants. Nat Protoc 1: 387–396 [DOI] [PubMed] [Google Scholar]
- Long SP, Humphries S, Falkowski PG. (1994) Photoinhibition of photosynthesis in nature. Annu Rev Plant Physiol 45: 633–662 [Google Scholar]
- Matt P, Schurr U, Klein D, Krapp A, Stitt M. (1998) Growth of tobacco in short-day conditions leads to high starch, low sugars, altered diurnal changes in the Nia transcript and low nitrate reductase activity, and inhibition of amino acid synthesis. Planta 207: 27–41 [DOI] [PubMed] [Google Scholar]
- Muñoz-Bertomeu J, Cascales-Miñana B, Mulet JM, Baroja-Fernández E, Pozueta-Romero J, Kuhn JM, Segura J, Ros R. (2009) Plastidial glyceraldehyde-3-phosphate dehydrogenase deficiency leads to altered root development and affects the sugar and amino acid balance in Arabidopsis. Plant Physiol 151: 541–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pärnik T, Keerberg O. (2007) Advanced radiogasometric method for the determination of the rates of photorespiratory and respiratory decarboxylations of primary and stored photosynthates under steady-state photosynthesis. Physiol Plant 129: 34–44 [Google Scholar]
- Porra RJ. (2002) The chequered history of the development and use of simultaneous equations for the accurate determination of chlorophylls a and b. Photosynth Res 73: 149–156 [DOI] [PubMed] [Google Scholar]
- Porra RJ, Thompson WA, Kriedemann PE. (1989) Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophyll a and chlorophyll b extracted with 4 different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim Biophys Acta 975: 384–394 [Google Scholar]
- Pruzinská A, Tanner G, Aubry S, Anders I, Moser S, Müller T, Ongania KH, Kräutler B, Youn JY, Liljegren SJ, et al. (2005) Chlorophyll breakdown in senescent Arabidopsis leaves: characterization of chlorophyll catabolites and of chlorophyll catabolic enzymes involved in the degreening reaction. Plant Physiol 139: 52–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reumann S, Weber AP. (2006) Plant peroxisomes respire in the light: some gaps of the photorespiratory C2 cycle have become filled—others remain. Biochim Biophys Acta 1763: 1496–1510 [DOI] [PubMed] [Google Scholar]
- Rontein D, Rhodes D, Hanson AD. (2003) Evidence from engineering that decarboxylation of free serine is the major source of ethanolamine moieties in plants. Plant Cell Physiol 44: 1185–1191 [DOI] [PubMed] [Google Scholar]
- Schreiber U, Schliwa U, Bilger W. (1986) Continuous recording of photochemical and nonphotochemical chlorophyll fluorescence quenching with a new type of modulation fluorometer. Photosynth Res 10: 51–62 [DOI] [PubMed] [Google Scholar]
- Schwacke R, Schneider A, van der Graaff E, Fischer K, Catoni E, Desimone M, Frommer WB, Flügge UI, Kunze R. (2003) ARAMEMNON, a novel database for Arabidopsis integral membrane proteins. Plant Physiol 131: 16–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarte S, Bauwe H. (2007) Identification of the photorespiratory 2-phosphoglycolate phosphatase, PGLP1, in Arabidopsis. Plant Physiol 144: 1580–1586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sienkiewicz-Porzucek A, Nunes-Nesi A, Sulpice R, Lisec J, Centeno DC, Carillo P, Leisse A, Urbanczyk-Wochniak E, Fernie AR. (2008) Mild reductions in mitochondrial citrate synthase activity result in a compromised nitrate assimilation and reduced leaf pigmentation but have no effect on photosynthetic performance or growth. Plant Physiol 147: 115–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson JP, Di Leo R, Dhanoa PK, Allan WL, Makhmoudova A, Clark SM, Hoover GJ, Mullen RT, Shelp BJ. (2008) Identification and characterization of a plastid-localized Arabidopsis glyoxylate reductase isoform: comparison with a cytosolic isoform and implications for cellular redox homeostasis and aldehyde detoxification. J Exp Bot 59: 2545–2554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville CR. (2001) An early Arabidopsis demonstration: resolving a few issues concerning photorespiration. Plant Physiol 125: 20–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaresan V, Springer P, Volpe T, Haward S, Jones JD, Dean C, Ma H, Martienssen R. (1995) Patterns of gene action in plant development revealed by enhancer trap and gene trap transposable elements. Genes Dev 9: 1797–1810 [DOI] [PubMed] [Google Scholar]
- Sweetlove LJ, Lytovchenko A, Morgan M, Nunes-Nesi A, Taylor NL, Baxter CJ, Eickmeier I, Fernie AR. (2006) Mitochondrial uncoupling protein is required for efficient photosynthesis. Proc Natl Acad Sci USA 103: 19587–19592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi S, Bauwe H, Badger M. (2007a) The photorespiratory pathway is important for the repair of photodamaged photosystem II under CO2 limiting conditions. Photosynth Res 91: 209 [Google Scholar]
- Takahashi S, Bauwe H, Badger M. (2007b) Impairment of the photorespiratory pathway accelerates photoinhibition of photosystem II by suppression of repair but not acceleration of damage processes in Arabidopsis. Plant Physiol 144: 487–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi S, Murata N. (2005) Interruption of the Calvin cycle inhibits the repair of photosystem II from photodamage. Biochim Biophys Acta 1708: 352–361 [DOI] [PubMed] [Google Scholar]
- Timm S, Nunes-Nesi A, Pärnik T, Morgenthal K, Wienkoop S, Keerberg O, Weckwerth W, Kleczkowski LA, Fernie AR, Bauwe H. (2008) A cytosolic pathway for the conversion of hydroxypyruvate to glycerate during photorespiration in Arabidopsis. Plant Cell 20: 2848–2859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voll LM, Jamai A, Renné P, Voll H, McClung CR, Weber APM. (2006) The photorespiratory Arabidopsis shm1 mutant is deficient in SHM1. Plant Physiol 140: 59–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu QB, Li G, Wang G, Sun JC, Wang PC, Wang C, Mi HL, Ma WM, Cui J, Cui YL, et al. (2008) Construction of a chloroplast protein interaction network and functional mining of photosynthetic proteins in Arabidopsis thaliana. Cell Res 18: 1007–1019 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Sun K, Sandoval FJ, Santiago K, Roje S. (2010) One-carbon metabolism in plants: characterization of a plastid serine hydroxymethyltransferase. Biochem J 430: 97–105 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.