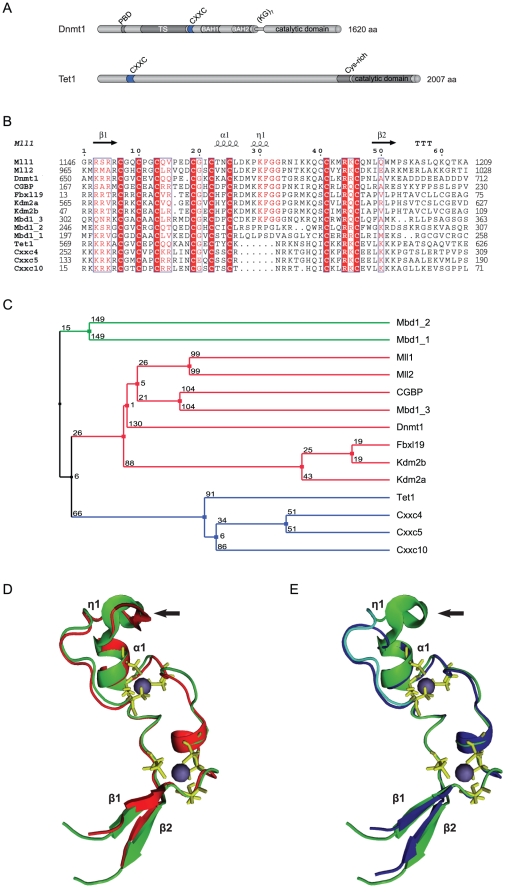
Figure 1. Sequence and predicted structural homology of CXXC domains.
(A) Schematic representation of the domain structure in Dnmt1 and Tet1. The catalytic domain and the N-terminal region of Dnmt1 are connected by seven lysine-glycine repeats [(KG)7]. PBD: PCNA binding domain; TS: targeting sequence; CXXC: CXXC-type zinc finger domain; BAH1 and 2: bromo-adjacent homology domain; NLS: nuclear localization signal; Cys-rich: cysteine rich region. (B) Alignment of mammalian CXXC domains. Numbers on the right side indicate the position of the last amino acid in the corresponding protein. The Mbd1a isoform contains three CXXC motifs (Mbd1_1-3). Absolutely conserved residues, including the eight cysteines involved in zinc ion coordination are highlighted in red and the conserved KFGG motif is in red bold face. Positions with residues in red face share 70% similarity as calculated with the Risler algorithm [66]. At the top residues of MLL1 involved in β sheets β1 and β2 (black arrows), α helices α1 and α2 and strict α turns (TTT) are indicated. All sequences are from M. musculus. Accession numbers (for GenBank unless otherwise stated): Dnmt1, NP_034196; Mll1, NP_001074518; Mll4, O08550 (SwissProt); CGBP, NP_083144; Kdm2a, NP_001001984; Kdm2b, NP_001003953; Fbxl19, NP_766336; Mbd1, NP_038622; CXXC4/Idax, NP_001004367; CXXC5, NP_598448; CXXC10 (see Materials and Methods). (C) A homology tree was generated from the alignment in (B). The three subgroups of CXXC domains identified are in different colors. Average distances between the sequences are indicated. (D–E) Homology models of the mouse Dnmt1 (D; red) and Tet1 (E; blue) CXXC domains superimposed to the CXXC domain of MLL1 (green; [35]). MLL1 residues that were described to contact DNA according to chemical shift measurements [35] are cyan in (E), while cysteines involved in coordination of the two zinc ions are yellow. Arrows point to the KFGG motif in MLL1 and Dnmt1. The locations of α helices and β sheets are indicated as in (B).