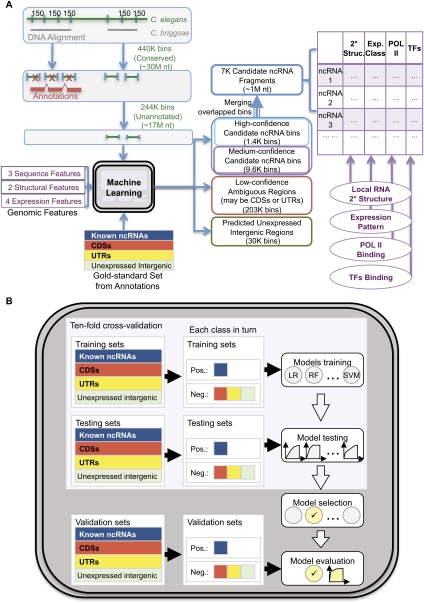
Figure 2.
A flowchart of incRNA (integrated ncRNA finder) for predicting and characterizing novel ncRNA candidates in C. elegans. (A) We looked for ncRNAs from conserved regions from the genome alignment between C. elegans and C. briggsae and divided them into small bins. Annotated bins from the gold-standard set were used to build a machine learning model based on nine expression, sequence, and structural features. The model was then used to score each unannotated bin by its likelihood of belonging to four genomic element classes (ncRNA, CDS, UTR, and unexpressed intergenic region). Adjacent bins predicted to be novel ncRNA candidates with high or medium confidence were merged into candidate ncRNA fragments, which were further characterized by their predicted RNA secondary structures, expression patterns, and the binding signals of Pol II and different transcription factors. (B) We used an unbiased procedure to build and evaluate our machine model. Multiple models were trained and tested using cross-validation, and the one with the highest cross-validation accuracy was evaluated using an independent validation set. For details, see Methods and Supplemental Methods.