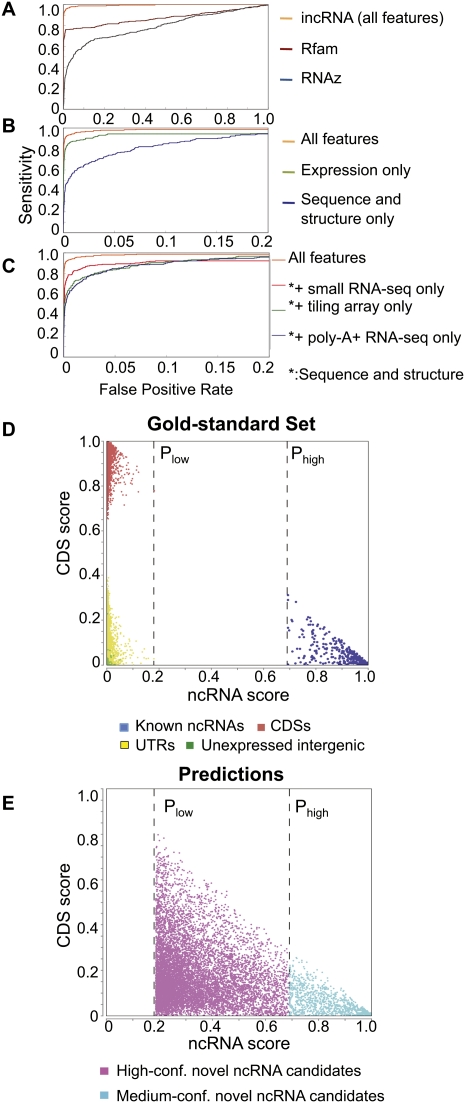
Figure 3.
Prediction performance of incRNA. (A) Comparison between the performance of incRNA and two previously published methods, Rfam/Infernal and RNAz. (B) Comparison between the performance of our method using all features, only expression data (tiling array and RNA-seq features), and sequence and structural features only (GC%, DNA conservation, RNA secondary structure, and protein conservation). (C) Comparison between the performance of our method using all features, and sequence and structural features in addition to only small RNA-seq, tiling array, or poly-A+ RNA-seq data. (D) Predicted ncRNA scores and CDS scores of annotated genomic elements (gold-standard set) assigned by our full model. All known ncRNA bins have ncRNA scores of at least Phigh and all other genomic elements (bins) have ncRNA scores of, at most, Plow. (E) Unannotated bins with ncRNA scores of at least Phigh form our high-confidence candidate ncRNA bins, while bins with ncRNA scores between Plow and Phigh form our medium-confidence candidate ncRNA bins. All predictions are applied on the pairwise alignments of C. elegans and C. briggsae.