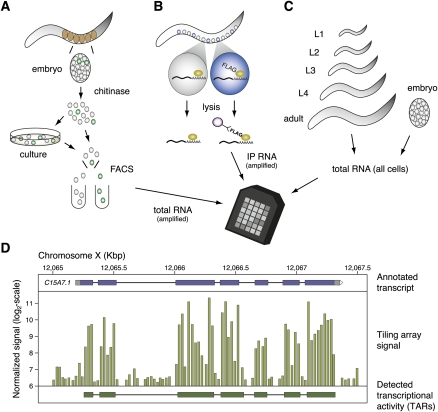
Figure 1.
Strategies for generating tiling array data sets from specific C. elegans cells in embryos and larvae and from whole animals at defined developmental stages. (A) In the MAPCeL (Micro-array Profiling of C. elegans Cells) method, embryos are isolated from gravid adults and blastomeres released by treatment with chitinase. Dissociated embryonic cells are either sorted immediately or cultured for 24 hrs before FACS. Total RNA is amplified for tiling array analysis. (B) The mRNA-tagging strategy was used to isolate RNA from specific larval and adult cells. The epitope-tagged (FLAG) polyA-binding protein (PAB-1) is expressed under the control of cell-specific promoters. The PAB-1:RNA complex is immunoprecipitated and RNA is amplified for tiling array analysis. (C) Total RNA is isolated from synchronized populations of embryonic, larval and adult animals for tiling array analysis. (D) Tiling array data (middle) is shown in a region around the annotated transcript C15A7.1 (top). Each vertical bar corresponds to the signal of one probe feature. A transcript identified by mSTAD using only the tiling array signal is shown at the bottom.