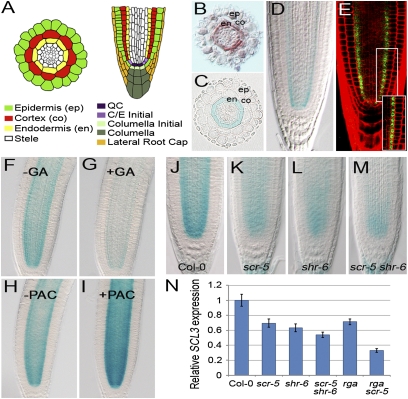
Fig. 1.
Transcriptional control of SCL3 in the root by the GA/DELLA and SHR/SCR pathways. (A) Schematic of the Arabidopsis root. (B–E) Localization of SCL3 mRNA and its protein. Expression of SCL3 is detected primarily in the endodermis by in situ hybridization (B) and a transcriptional fusion (pSCL3::GUS) (C and D). Blue staining is observed in a manner similar to the SCL3 in situ hybridization pattern. (E) Translational fusion (pSCR::SCL3-GFP) in the root. GFP signals are observed in the nuclei of the QC and the endodermis lineage. (F and G) pSCL3::GUS in WT in the absence (F) or presence (G) of 10 μM of exogenous GA3 for 6 h. (H and I) pSCL3::GUS in WT in the absence (H) or presence (I) of 10 μM of PAC for 6 h. Expression of SCL3 is modulated by bioactive GA contents. (J–M) Expression of pSCL3::GUS in Col-0 (J), scr-5 (K), shr-6 (L), and scr-5 shr-6 (M). (N) qRT-PCR of SCL3 transcripts in Col-0, scr, shr, scr shr, rga, and rga scr roots. The SCL3 transcript level is substantially reduced in rga scr double mutants. The SCL3 mRNA level in Col-0 is arbitrarily set to 1. Error bars indicate SD from three biological replicates.