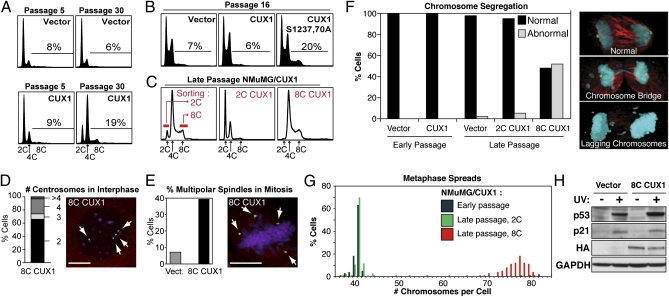
Fig. 1.
Tetraploidy and chromosomal instability in p110 CUX1-expressing NMuMG cells. (A) NMuMG cells stably expressing p110 CUX1 (amino acids 747–1505) were generated by retroviral infection. More than 500 colonies were pooled after selection, which was considered passage 1. Cell cycle distribution was determined by FACS. (B) FACS analysis was performed on independent populations of NMuMG cells expressing either p110 CUX1 or p110S1237,1270A CUX1. (C) Late passage NMuMG/CUX1 cells were FACS-sorted according to DNA content after Hoechst 33342 staining and put back in culture. 2C represent G1 diploid cells and 8C are G2/M tetraploids. (D and E) Number of centrosomes during interphase (D, n = 304) or spindle poles during early mitosis (E, n = 250 for each cell line) was determined by indirect immunofluorescence according to γ-tubulin staining (green), whereas microtubules and DNA were revealed by α-tubulin (red) and DAPI (purple) staining, respectively. (Scale bars, 10 μm.) (F) Chromosome segregation during anaphase was studied by confocal microscopy for late-passage NMuMG/vector (n = 31), early-passage NMuMG/CUX1 (n = 50), and late-passage 2C-sorted (n = 57) 8C-sorted (n = 23) NMuMG/CUX1 cells. Cells were stained for γ-tubulin (green), α-tubulin (red), and DNA (DAPI, blue). Images are a composite of z-sections encompassing entire cells. (G) Chromosome counts were determined on metaphase spreads prepared from early-passage NMuMG/p110 CUX1 or late-passage 2C- or 8C-sorted NMuMG/p110 CUX1 cells (n ≥ 100 each). (H) NMuMG/vector and 8C NMuMG/CUX1 cells were left untreated or exposed to 20 mJ/cm2 UV. Cell lysates were prepared 24 h later and analyzed by SDS/PAGE and Western blotting using the indicated antibodies.