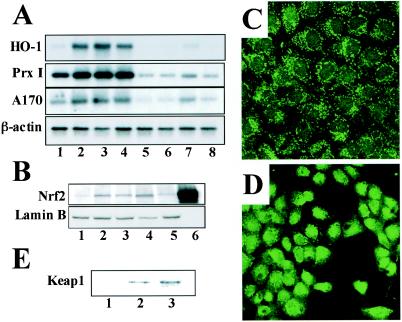
FIG. 4.
cyPGs activate Nrf2 in macrophages and hepatocytes. (A) Peritoneal macrophages derived from either wild-type mice (lanes 1 to 4) or nrf2−/− mice (lanes 5 to 8) were treated with 5 μM (lanes 2 and 6) or 10 μM (lanes 3 and 7) 15d-PGJ2 or with 100 μM diethylmaleate (lanes 4 and 8). Untreated controls are shown in lanes 1 and 5. Total RNAs were analyzed by RNA blotting analysis, using cDNAs for HO-1, PrxI, or A170 as probes. A β-actin cDNA probe was used for a loading control. (B) Nuclear extracts were prepared from macrophages untreated (lane 1) or treated with diethylmaleate (100 μM) (lane 2), 15d-PGJ2 (5 μM) (lane 3), PGA1 (100 μM) (lane 4), or PGE2 (100 μM) (lane 5). The extracts were immunoblotted with anti-Nrf2 antibody or anti-lamin B antibody. Recombinant Nrf2 in 293T cells was also loaded as a control (lane 6). (C and D) RL34 cells were either untreated (C) or treated with 10 μM 15d-PGJ2 for 4 h (D), and expression of Nrf2 were examined immunocytochemically with the anti-Nrf2 antibody. Subcellular localization of Nrf2 was analyzed by confocal microscopy. (E) RL34 cells were treated with dimethyl sulfoxide (lane 1), biotinylated Δ12-PGJ2 (lane 2), or biotinylated 15d-PGJ2 (lane 3). Keap1-PGJ2 complexes were precipitated from the cell extracts with avidin beads and probed with the anti-Keap1 antibody.