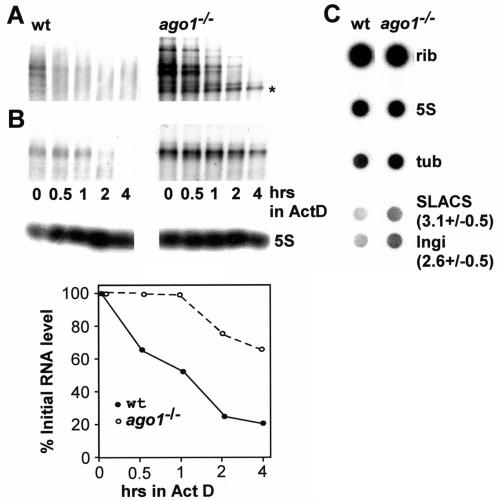
FIG. 7.
AGO1 affects the accumulation of Ingi and SLACS transcripts. (A) Northern blot of Ingi transcripts in wild-type (wt) and ago1−/− cells over a period of 4 h following the addition of actinomycin D. The asterisk indicates a transcript which appeared to be more long lived in ago1−/− cells than in wild-type cells. (B) Northern blot of SLACS transcripts in wild-type and ago1−/− cells over a period of 4 h following the addition of actinomycin D (upper panel). Hybridization to 5S RNA served as a loading control (middle panel). SLACS transcripts were quantitated relative to 5S RNA (lower panel). (C) Newly synthesized RNA from wild-type and ago1−/− cells was hybridized to the following DNAs spotted onto a nitrocellulose filter: large rRNAs (rib), 5S rRNA (5S), α-tubulin (tub), SLACS, and Ingi. The numbers next to the SLACS and Ingi hybridizations indicate the fold increase in ago1−/− cells relative to wild-type cells. The experiment was repeated three times, and the standard deviation is indicated.