FIG. 5.
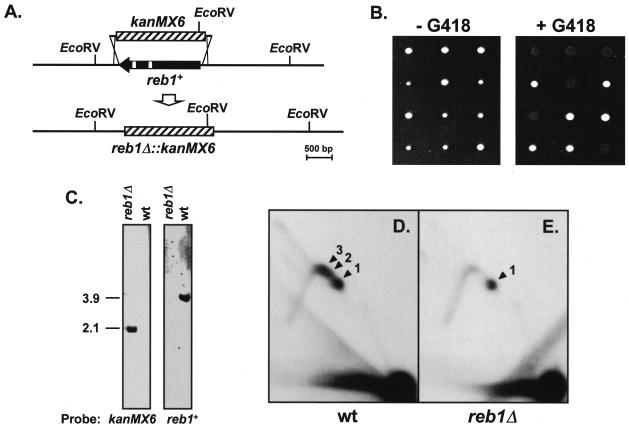
Deletion of the reb1+ gene. (A) Diagrammatic representation of the reb1+ gene deletion by PCR-mediated disruption. One allele of the reb1+/reb1+ diploid strain 117×118 was replaced by the G418-resistant gene kanMX6+ (hatched box). Open small boxes within reb1+ represent introns. The locations of two EcoRV restriction sites flanking reb1+ and an additional one within kanMX6+ are indicated. (B) Tetrad analysis of three selected heterozygous diploids. Spores from three asci were separated and grown in yeast extract with supplements without G418 (left panel). Colonies were then replicated onto a new plate containing 100 μg of G418/ml (right panel) to determine the segregation of the alleles. (C) Southern blot verifying the deletion of reb1+ gene by replacement with kanMX6+. EcoRV-digested DNA from reb1Δ and wild-type haploid cells was hybridized with a probe specific for kanMX6+ (left panel) or reb1+ (right panel). The kanMX6+ probe hybridized to the expected 2.1-kb restriction fragment from reb1Δ DNA, containing most of the kanMX6+ gene (see bottom map in panel A) and did not hybridize to the DNA from wild-type cells. As expected, the reb1+ probe did not hybridize to the DNA from reb1Δ cells but detected the 3.9-kb fragment containing reb1+ in the DNA from wild-type cells. (D and E) 2D gels of plasmid pIRT1.6(+) replicating in wild-type (wt) and reb1Δ cells, respectively. The restriction fragment analyzed was the same as in Fig. 2C. Arrowheads labeled 1 to 3 point to the spots of accumulated replication intermediates induced by the three barriers in wt cells. Note that in reb1Δ cells only the spot generated by RFB1 remained. The probe used in these autoradiograms was the same as for Fig. 2.