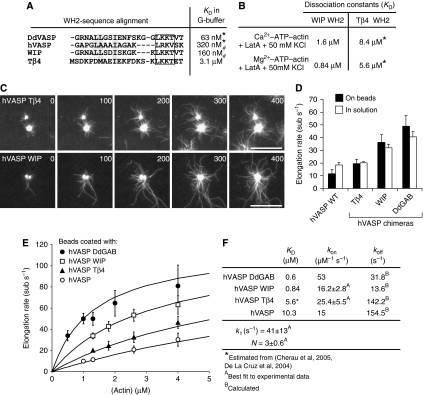
Figure 4.
Analysis of hVASP WH2 chimeras reveals VASP-mediated filament elongation is enhanced by saturable monomer binding to GAB sites. (A) Sequence alignment of the WH2 motifs is indicated. In the chimeric proteins, the GAB of hVASP was replaced by the WH2 motifs from Tβ4 and WIP. KD values of the actin–WH2 interaction in G-buffer where either determined in this study (*) or by Chereau et al (2005) (#). Conserved hydrophobic residues are highlighted in grey and the conserved LxxV/T motifs (x=basic amino acid) are boxed. (B) KD values determined as for Figure 3A and B under the conditions are indicated. (*) The KD values for the Tβ4–WH2 interaction were estimated on the basis of previous studies (De La Cruz et al, 2000; Hertzog et al, 2002; Chereau et al, 2005). (C) TIRFM micrographs of the assembly of 1.3 μM actin (30% OG labelled) on beads saturated with hVASP Tβ4 and hVASP WIP in TIRF buffer in the presence of 80 nM CP. Both chimeras processively elongate actin filaments. Time is indicated in seconds, scale=20 μm. (D) Elongation rates mediated by hVASP and the chimeras hVASP Tβ4, hVASP WIP and hVASP DdGAB at 1.3 μM G-actin either with 500 nM of the VASP constructs in solution or in the presence of 80 nM CP on saturated beads. Number of analysed filaments >20 for bead assays and >40 for assays in solution. Elongation rates are presented as mean values±s.e.m. (E) Elongation rates obtained from TIRF assays with beads coated with different hVASP constructs in the presence of 80 nM CP at the actin concentrations indicated. Solid lines represent best fits of the experimental data using the mathematical model for processive filament elongation by immobilized VASP as described in Figure 5 and the Materials and methods section. Elongation rates are presented as mean values±s.e.m. (F) Parameters derived from fitting of the data are shown in (E).