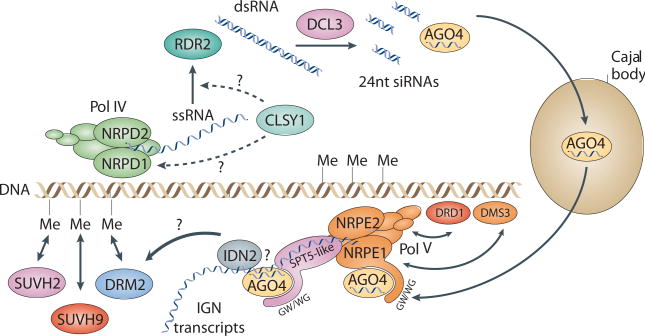
Figure 2. Model for RNA directed DNA methylation (RdDM).
Single-stranded RNA transcripts (ssRNA) corresponding to transposons and repeat elements are hypothesized to be generated by Pol IV. CLSY1, a putative chromatin remodeling factor, likely functions early in RdDM, possibly recruiting Pol IV to chromatin or aiding in ssRNA transcript processing. RDR2, an RNA dependant RNA polymerase, is proposed to generate double-stranded RNA (dsRNA) from the ssRNA transcripts. DCL3, a dicer-like protein, is thought to process the dsRNAs into 24nt siRNAs, which are bound by an argonaute protein, AGO4. AGO4 localizes to Cajal bodies and while the function of this association remains unknown, it appears necessary for wild-type levels of RdDM33. AGO4 also co-localizes with two Pol V subunits, NRPE1 and NRPE2, and DRM2 at a distinct nuclear foci, the AGO4/NRPD1b(NRPE1) body (not depicted), that may represent a site of active RdDM33. Pol V is thought to transcribe intergenic noncoding (IGN) regions throughout the genome. NRPE1 association with chromatin requires another putative chromatin remodeling factor, DRD1, and an SMC domain protein, DMS3. IGN transcripts may serve as a scaffold to recruit AGO4, which interacts with the GW/WG motifs of NRPE1 and SPT5-like, possibly through interactions between AGO4-bound siRNAs and the nascent transcript. An RNA binding protein, IDN2, is proposed to recognize the siRNA/nascent transcript duplex. These associations may aid in targeting DRM2 to genomic loci that produce both 24nt siRNAs and IGN transcripts. Recruitment or retention of DRM2 at such loci may be aided by SUVH9 and SUVH2, two proteins that bind methylated DNA and likely act late in RdDM. Me, DNA methylation.