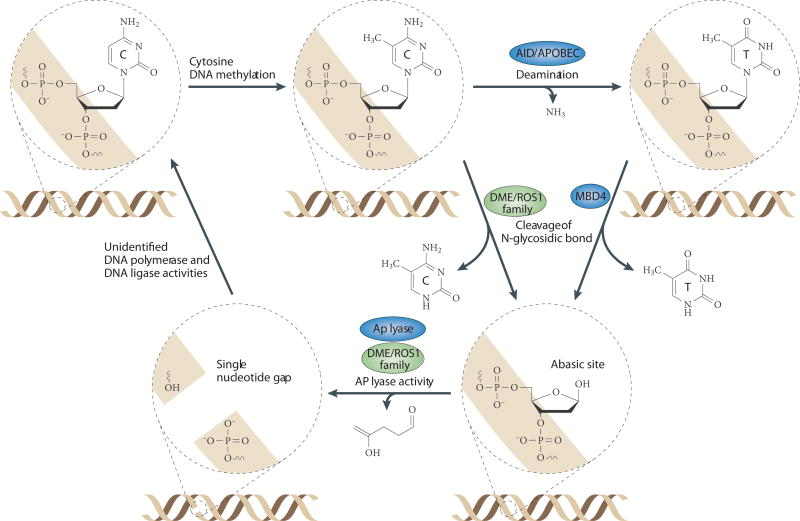
Figure 7. Active DNA Demethylation via DNA glycosylase activity and Base Excision Repair.
In Arabidopsis (green shaded proteins) methylated (CH3) cytosine (C) bases are removed by the DEMETER (DME)/ROS1 family of bifunctional 5-methylcytosine glycosylases. First, the methylated cytosine base is released by cleavage of the N-glycosidic bond, generating an abasic site. Next the phosphodiester linkage is broken both 3’ and 5’ of the abasic site through apyrimidic (AP) lysase activity, generating a single nucleotide gap in the DNA. The DNA is then proposed to be repaired by unknown DNA polymerase and ligase activities, resulting in a net loss of cytosine methylation. In Zebrafish and mammals, no efficient 5-methylcytosine glycoslases have been identified. However, in Zebrafish (blue shaded proteins), it has been proposed that the AID and APOBEC family of deaminases first convert methylated cytosines into thymines (T), generating thymine/guanine (G) (T:G) mismatches. Then, these mismatches could be recognized by the MBD4 glycosylase, resulting in removal of the thymine base and generation of an abasic site. Unlike the DME/ROS1 glycosylases, MBD4 is a monfunctional DNA glycosylase, thus another unidentified protein is likely required to provide the AP lyase activity in order to remove the sugar ring to generate a single nucleotide gap. As in Arabidopsis, this substrate is proposed to be repaired by unidentified DNA polymerase and ligase activities.