Fig. 3.
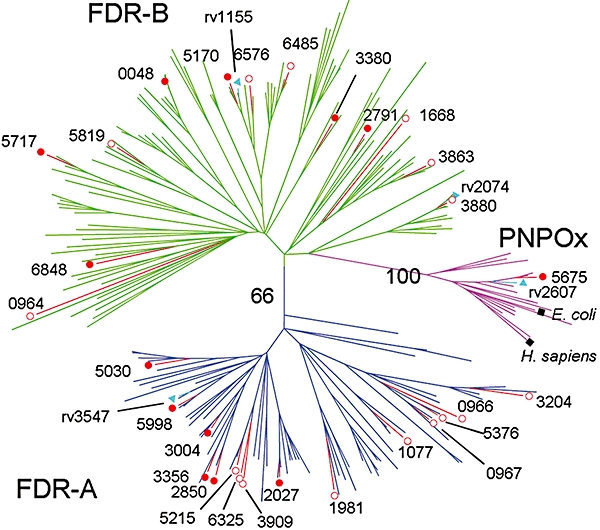
Phylogenetic relationship between the PNPOx, FDR-A and FDR-B families. A condensed tree was constructed from 146 protein sequences of FDRs/PNPOxs from seven species of Actinomycetales (M. smegmatis, M. tuberculosis H37Rv, M. vanbaalenii, Rhodococcus sp. RH1, Arthrobacter sp. FB24, S. coelicolor, Frankia alni and Nocardioides sp. JS614), plus the known PNPOx enzymes from H. sapiens, E. coli, Saccharomyces cerevisiae, Caenorhabditis elegans and Mus musculus. Solid red circles denote the FDRs described in this paper, and open red circles denote other potential FDRs from M. smegmatis (TIGR locus number shown). For clarity, only enzymes from other species that have been previously characterized are labelled: blue triangles denote M. tuberculosis enzymes with known structures or previously described functions and black squares denote previously described PNPOxs.
