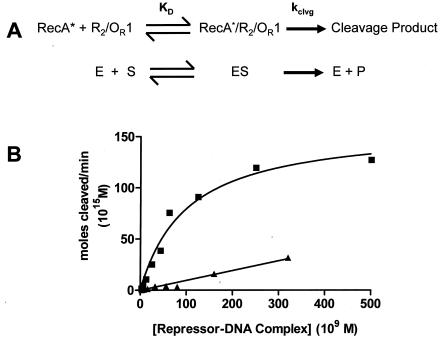
FIG. 6.
Kinetic analysis of RecA-mediated autoproteolysis of 434 repressor-DNA complexes. (A) Diagrammatic comparison of the similarities between RecA-mediated repressor autocleavage and the enzyme-catalyzed Michaelis-Menton reaction. (B) Maximum velocity of repressor autocleavage at a fixed concentration of repressor (as determined in Fig. 5) plotted as a function of the concentration of repressor-OR1 (▪) and repressor-OR2 (▴) complexes. The lines represent fits to the Michaelis-Menten equation. The fitting parameters are given in the text. KD, dissociation constant; E, enzyme; S, substrate; ES, enzyme-substrate complex; P, product.