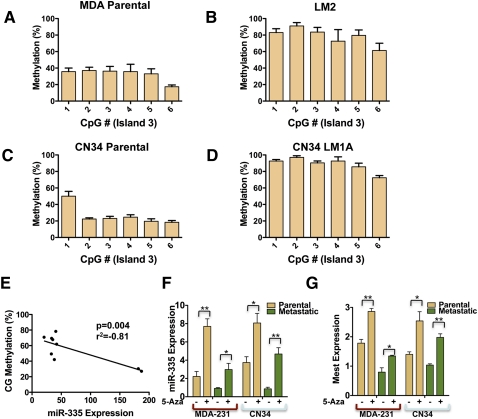
Figure 3.
Pyrosequencing reveals the methylation status of CpG island 3 as a predictor of miR-335 expression, and inhibition of DNA methylation is sufficient to restore miR-335 expression in metastatic breast cancer cells. (A–D) Pyrosequencing of bisulfite-treated DNA reveals the methylation percentage (Y-axis) as a function of CpG dinucleotide position in island 3 of poorly metastatic breast cancer populations (MDA parental and CN34 parental) and their highly metastatic derivatives (LM2 and CN34LM1A). All error bars represent SEM. (E) CpG methylation percentage as a function of miR-335 expression (correlation coefficient r2 = −0.81; P = 0.004). (F) Quantitative real-time PCR expression of miR-335 expression in parental MDA-231 breast cancer line and its metastatic LM2 derivative and the CN34 primary malignant population and its metastatic derivative (CNLM1A) in the presence or absence of 5-Aza (5 μM treatment for 96 h for MDA lines and 1 μM for CN34 lines; metastatic lines, n = 6; parental lines, n = 3). P-values represent unpaired one-tailed t-test significance values. (*) P < 0.05; (**) P < 0.005. (G) Quantitative real-time PCR expression of Mest expression in parental MDA-231 breast cancer line and metastatic LM2 derivative in the presence or absence of 5-Azacytidine as in F; (n = 3). (*) P < 0.05; (**) P < 0.005.