Abstract
Design templates that involve discovery, analysis, and integration of information resources commonly occur in many scientific research projects. In this paper we present examples of design templates from the biomedical translational research domain and discuss the requirements imposed on Grid middleware infrastructures by them. Using caGrid, which is a Grid middleware system based on the model driven architecture (MDA) and the service oriented architecture (SOA) paradigms, as a starting point, we discuss architecture directions for MDA and SOA based systems like caGrid to support common design templates.
1. Introduction
Scientific research in any field encompasses a wide range of problems and application scenarios. While a variety of approaches are developed by researchers to attack specific sets of problems, common aspects of these approaches can be grouped into domain-specific general patterns of research. We refer to such patterns as design templates. A design template is a representation of the common understanding of a domain problem by researchers. It describes the common components of the problem and generic approaches to attacking the problem. The importance of design templates is that they capture design requirements and constraints that arise in broad families of applications
We focus on translational biomedical research design templates. The term “translational biomedical research” is associated with research directed at developing ways of treating or preventing diseases through the application of basic science knowledge or techniques. Translational research projects are heterogeneous in nature. They target a wide variety of diseases, test many different kinds of biomedical hypotheses, and employ a large assortment of experimental methodologies. Specific translational research problems are often used as motivating use cases for computer science research. We will present an initial set of “design templates” that capture the salient aspects of different types of translational research studies. In this paper we relate translational research design templates to middleware requirements. This work is motivated both by Christopher Alexander’s seminal writings on design languages used to capture and classify salient aspects of architectural design [1] as well as the later work on software design patterns [2] where somewhat analogous principles were applied to software design.
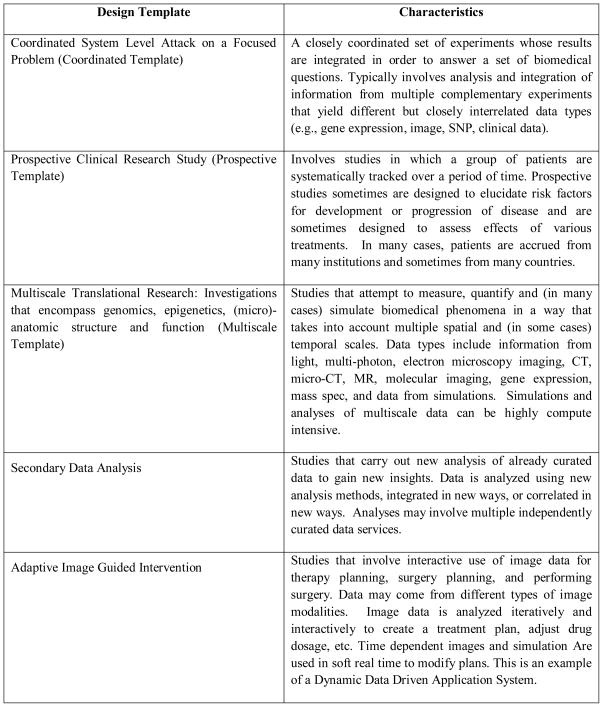
Note that different aspects of a given real world translational research study can often be described by more than one design template. Figure 1 shows several examples of design templates in translational research and their primary characteristics.
Figure 1.
Examples of design templates for translational research.
We describe middleware requirements arising from the first three design templates listed in Figure 1; 1) coordinated system level attack on focused problem, 2) prospective research studies and 3) multiscale translational research: investigations that encompass genomics, epigenetics, (micro)-anatomic structure and function.
Discovery, analysis, and integration of heterogeneous information resources is a theme that commonly occurs in many translational research design templates. In recent years, the service oriented architecture (SOA) and the model driven architecture (MDA) have gained popularity as frameworks on which to develop interoperable systems. The service-oriented architecture (SOA) encapsulates standards (e.g., Web Services[3], Web Services Resource Framework[4]) on common interface syntax, communication and service invocation protocols, and the core capabilities of services. The model driven architecture promotes a software design approach based on platform independent models and metadata to describe these models. Solutions integrating and extending these architecture patterns offer a viable approach to address the problem of programmatic, syntactic, semantic interoperability, and integration, and as a consequence the implementation of design templates. Using caGrid[5, 6], which combines and extends MDA and SOA for scientific research, as a starting point, we discuss architecture features and tools for caGrid like systems to address the requirements of design templates in scientific research.
2. Examples of Design Templates
In this section we will describe the following three design templates: 1) Coordinated Systems Level Attack on Focused Problem (Coordinated Template), 2) Prospective clinical research study (Prospective Template) and 3) Multiscale Translational Research: Investigations that encompass genomics, epigenetics, (micro)-anatomic structure and function (Multiscale Template). The other three design templates are outlined in Figure 1; we will expand on those elsewhere. In conjunction with the design templates, we also present requirements that arise in these templates.
The Coordinated Template involves a closely coordinated set of experiments whose results are integrated in order to answer a set of biomedical questions. A good example of an application described by this design template is the effort on the part of the Cardiovascular Research Grid (CVRG) and the Reynolds project to integrate genomic, proteomic, ECG and image data to better predict the likelihood of potentially lethal arrhythmias (http://www.cvrgrid.org). This problem is of great practical significance because at risk patients can receive implantable cardioverter defibrillators (ICDs). Another example of this design template is the effort on the part of the NCI ICBP funded Ohio State Center for Epigenetics (http://icbp.med.ohio-state.edu) to understand the impact of epigenetic changes on particular genomic pathways through coordinated study of gene epigenetics, gene sequence, gene expression, and proteomics. A deep understanding of this integrated system can be used to develop new drugs and to evaluate which patients are best suited for a given therapy.
The Coordinated Template provides motivating requirements for the development of methods for supporting deep semantic integration of many complementary types of information. Gene sequence, genetic expression, epigenetics, and protein production need to be interpreted, represented, and modeled as highly interdependent phenomena in this design template. In the CVRG example, researchers access different data systems to create candidate patient profiles using clinical, genomic, proteomic, ECG data, and information derived from images. These candidate patient profiles are being compared and analyzed by CVRG researchers to predict likelihood of potentially lethal arrhythmias.
The Prospective Template involves studies in which a group of patients are systematically followed over a period of time. Prospective studies sometimes are designed to elucidate risk factors for development or progression of disease and are sometimes designed to assess effects of various treatments. In many cases, patients are accrued from many institutions and sometimes from many countries.
The Prospective Template provides motivating requirements for security, semantic interoperability, and interfacing with existing institutional systems. It is very expensive and (with a few exceptions) impractical to develop a purpose built information system for a particular prospective study. From the point of view of economics, logistics, and quality control, it makes much more sense to share a core information architecture for many prospective trials. Prospective template information architectures need to interoperate with existing institutional systems in order to better support prospective trial workflow, and to avoid double entering of data and manual copying of files arising from Radiology and Pathology. Prospective trials have a huge semantic scope; there is a vast span of possible diseases, treatments, symptoms, Radiology and Pathology findings, genetic and molecular studies. There is a need to translate between ontologies, controlled vocabularies, and data types. Subsystems that need to be interacted with may be commercial or open source systems that adhere to varying degrees to a broad collection of distinct but overlapping standards including HL7 (www.hl7.org), DICOM (http://medical.nema.org/), IHE (http://www.ihe.net/), LOINC (http://www.nlm.nih.gov/research/umls/loinc_main.html), caBIG™ Silver and Gold [7].
There are a variety of efforts to address the issue of integrating information across trials. These include the Clinical Data Interchange Standards Consortium (CDISC) and the HL7 based Regulated Clinical Research Information Management (RCRIM) Technical Committee (TC). The Biomedical Research Integrated Domain Group (BRIDG) project described in [8] aims to systematically harmonize existing clinical research standards and to systematically develop specifications for new standards to support clinical research.
The Multiscale Template models studies that attempt to measure, quantify, and in some cases simulate, biomedical phenomena in a way that takes into account multiple spatial and/or temporal scales. The study of tumor microenvironment is one excellent example. The development of cancer occurs in both space and time. Cancers are composed of multiple different interacting cell types; the genetics, epigenetics, regulation, protein expression, signaling, growth and blood vessel recruitment take place in time and space. Very large scale datasets are required to describe tumor microenvironment experimental results. Tumor microenvironment datasets are semantically complex as they encode ways in which morphology interrelates over time with genetics, genomics, epigenetics, and protein expression.
Image acquisition, processing, classification and analysis play a central role in support of the Multiscale Template. A single high resolution image from digitizing microscopes can reach tens of gigabytes in size. Hundreds of images can be obtained from one tissue specimen, thus generating both 2D and 3D morphological information. In addition, image sets can be captured at multiple time points to form a temporal view of morphological changes. Images obtained from queries into image databases are processed through a series of simple and complex operations expressed as a data analysis workflow. The workflow may include a network of operations such as cropping, correction of various data acquisition artifacts, filtering operations, segmentation, registration, feature detection, feature classification, as well as interactive inspection and annotation of images. The results of the analysis workflow are annotations on images and image regions, which represent cells (a high resolution image may contain thousands of cells), cell types, and the spatial characteristics of the cells. These annotations may be associated with concepts and properties defined in different ontologies. The researcher may compose queries to select a subset of images with particular features (e.g., based on cell types and distribution of cells in the image) and associate these image features with genomic data obtained for different types of cells. Genetic and cellular information can further be integrated with biological pathway information to study how genetic, epigenetic, and cellular changes may impact major pathways.
Simulation also plays a crucial role in the Multiscale Template. As knowledge of basic biomedical phenomena increases, the ability to carry out meaningful detailed simulations dramatically increases. Some researchers are now carrying out tumor microenvironment simulations[9] and we expect the prevalence of this to dramatically increase with the improved quality of detailed multiscale data.
3. Middleware Architecture Features
In this section we will look at architecture features for middleware systems as motivated by the translational research design templates in a Grid environment. We focus on five core areas: management of data structures and semantic information, support for data and analytical services, support for federated query and orchestration of services, interoperability with other infrastructure, and security infrastructure. We will use caGrid [5, 6, 10] along with references to several other systems to show implementation choices in these areas. The key architecture features of caGrid are based on a wide range of design templates from translational research and other biomedical research areas targeted by the cancer Biomedical Informatics Grid (caBIG™) program (https://cabig.nci.nih.gov). A key feature of caGrid is that it draws from the basic principles and marriage of the model driven architecture (MDA) and the service oriented architecture (SOA) and extends them with the notion of strongly typed services, common data elements, and controlled vocabularies to address the design templates. As such, caGrid is a good example of how middleware systems combining MDA and SOA can address the requirements of design templates and is a good starting point to describe ideas as to what additional capabilities are needed in those systems.
3.1. Management of Data Structures and Semantic Information
The three design template examples require semantic interoperability and integration of information from multiple data types and data sources. Successful implementation in a multi-institutional setting of these design templates is impacted by 1) how effectively the researcher can discover information that is available and relevant to the research project and 2) how efficiently he can query, analyze, and integrate information from different resources. One obstacle is the fact that distributed data sources are oftentimes fragmented and not interoperable. Datasets vary in size, type, and format and are managed by different types of database systems. Naming schemes, taxonomies, and metadata used to represent the structure and content of the data are heterogeneous and managed in silos; any two databases may define data that contain the same content with completely different structure and semantic information.
In order for any two entities to correctly interact with each other (a client program with a resource, or a resource with another resource), they should agree on both the structure of and the semantic information associated with data object(s) they exchange. An agreement on the data structure is needed so that the program consuming an object produced by the other program can correctly parse the data object. Agreement on the semantic information is necessary so that the consumer can interpret the contents of the data object correctly. In most Grid projects, however, data structures and semantic information are represented and shared in a rather ad hoc way; they are maintained in silos or embedded deep in the middleware code or application logic.
caGrid addresses this problem as follows in the caBIG™ program; this support in the current caGrid 1.0 architecture has been described in [5, 10], we provide a summary of that description here. Each data or analytical resource is made accessible through application programming interfaces (APIs) that represent an object-oriented view of the resource. The APIs of a resource operate on published domain object models, which are specified as object classes and relationships between the classes. caGrid leverages several data modeling systems to manage and employ these domain models. Data types are defined in UML and converted into ISO/IEC 11179 Administered Components. These components are registered in the Cancer Data Standards Repository (caDSR)[11] as common data elements. The definitions of the components are based on vocabulary registered in the Enterprise Vocabulary Services (EVS)[11]. In this way, the meaning of each data element and relationships between data elements are described semantically. At the grid level, objects conforming to registered domain object models are exchanged between Grid end points in the form of XML documents, i.e., an object is serialized into an XML document to transfer it over the wire. caGrid adopts the strongly-typed service paradigm in that an object is serialized into an XML document that conforms to an XML schema registered in the Mobius GME service[12]. Thus the XML structure of the object is available to any client or resource in the environment. In short, the properties and semantics of data types are defined in caDSR and EVS and the structure of their XML materialization in the Mobius GME.
The curation and publication of semantically annotated common data elements in caBIG is done through a review process that allows freedom of expression from data and tool providers, while still building on a common ontological backbone[7]. This model has worked relatively well in the controlled environment of the three year pilot phase of the program. However, as the number of caBIG participants, projects, and tools continues to grow, a number of stress points are arising for the program and its infrastructure. Probably the most notable such point is the reliance on community review, guidance, and curation of every single data type used on the Grid. Achieving the highest level of interoperability, Gold compliance, requires among other things, that a service’s data model is reviewed, harmonized, and registered [7]. These efforts result in the identification of common data elements, controlled vocabularies, and object-based abstractions for all cancer research domains. This model is both the heart of the success of the caBIG approach and its biggest obstacle for would-be adopters or data and tool providers. Projects and communities, for which the existing model is likely to incur high development costs, are those working in new domains with data types and concepts that may be partially or largely uncovered by the existing ontology and data models. Such projects must go through the process of harmonizing and registering all missing models and ontologies into the environment. This can be a daunting task, and scaling the infrastructure and processes to accommodate such communities will be critical to its success. A tempting view point is to simply allow these projects to either not register their models or anchor them to the shared ontology. This however, results in a fundamental break down in the approach, and creates the very silos of non-interoperable applications which caBIG and caGrid set out to integrate. Maintaining the high level of integrity necessary for an ontology backbone without centralized control will be a key challenge, as well as a necessity to scale towards the next generation of science grid systems. The caBIG community and the caGrid development effort are starting to investigate support for approaches towards federating the storage, management, and curation of terminologies in the larger distributed environment, while facilitating and promoting harmonization with a common ontological backbone and reuse of community accepted standard data elements and terminologies.
Grid middleware systems that employ SOA make use of XML for service interface descriptions, service invocations, and data transfers. A service’s interface is described with Web Services Description Language (WSDL). Data is exchanged between two Grid end points in structured XML messages. In the case of caGrid, service interfaces are described in WSDL conforming to the Web Services Resource Framework standards. Data objects are serialized into structured XML documents conforming to registered XML schemas. The objects correspond to instances of classes in an advertised logical object-oriented model and the semantics of the data is described through annotations of this model. Both the model and the annotations are derived from the aforementioned curated data types. It is through this formal connection between the XML Schemas and the curated and annotated data models, that the semantic meaning of the structured XML can be understood. That is, there is a formal binding between the structure of the data exchanged, and its underlying semantic meaning. Current work is underway in caGrid to better surface the query, access, and management of this binding on the Grid itself. Similarly, the utility of more directly annotating the XML layer with semantic information, as opposed to making it indirectly accessible through formal mappings, is being investigated. For example, the W3C currently has a recommendation for Semantic Annotations for WSDL and XML Schema (http://www.w3.org/TR/sawsdl/). It defines a set of extension attributes for the Web Services Description Language (WSDL) and XML Schema definition language that allows description of additional semantics of WSDL components. SAWSDL does not specify a language for representing the semantic models, but instead provides mechanisms by which concepts from the semantic models can be referenced from within WSDL and XML Schema components using annotations. This is particularly attractive, as it provides a framework to directly annotate corresponding artifacts, without requiring any particular constraints on our existing semantic infrastructure.
Similar approaches are being investigated for dealing with prohibitively large, or natively binary, data in caGrid. Some data types relevant to the cancer community are not easily, or at least efficiently, represented as XML. Examples include microarray experiment results, which are generally massive numbers of floating point numbers, and images encoded in the industry standard Digital Imaging and Communications in Medicine (DICOM) format. One issue is the efficient transfer of large scale data between Grid end points (between clients and services or between two services). The caGrid infrastructure provides service components, one component being implemented using Java (for portability across platforms) and one component using GridFTP[13, 14], for bulk transfer of data in binary format. The other issue is how to represent the structure of the data being transferred in binary format. caGrid is currently investigating work such as the Open Grid Forum’s Data Format Description Language WG (DFDL-WG). The aim of this working group is to define an XML-based language, the Data Format Description Language (DFDL), for describing the structure of binary and character encoded (ASCII/Unicode) files and data streams so that their format, structure, and metadata can be exposed. This work allows a natively binary or ASCII data set to be formally described by an annotated XML schema in such away as it can be processed transparently in either its native format, or as an XML document (with the parser performing the necessary translations). Coupled with XML schema associated semantic annotations, this would allow semantic interoperability of binary data and seamless exchange on the Grid.
3.2. Data and Analytical Services
In practice, it is reasonable to assume that studies conforming to the example design templates will involve access to databases and analytical methods supported by heterogeneous systems. The Coordinated Template, for example, may involve datasets that are stored in a combination of relational database management systems (e.g., SNP data in the CVRG example) and XML database management systems (e.g., the ECG data in the CVRG example). Analytical programs may have been implemented on a variety of platforms (e.g., Matlab, C++, and Java). In most cases, it is prohibitively expensive or infeasible (due to security and ownership concerns) to copy data to a single DBMS or entirely port the implementation of an analytical program. It is also not efficient to develop clients that have specific modules for each different database system or analytical program.
The MDA and the SOA facilitates system-level interoperability by standardization of data exchange protocols, interface representations, and invocation mechanisms in a Grid environment. They, however, also introduce new requirements on resources to join a Grid environment: resources are exposed as (object-oriented) services; these services export well-defined and strongly-typed interfaces; and rich metadata is associated with each service and advertised in the environment. In order for Grid middleware architectures to be successful in scientific domain, the impact of these additional requirements as a barrier to entry of resources to the Grid environment should be minimized. Additional capabilities are needed to efficiently support development of Grid nodes, which can leverage MDA and SOA together and can utilize common authentication and authorization.
The Prospective Template is a particularly strong driver for the need for standardizations on protocols, infrastructure of support services (such as Grid-wide management of structure and semantics of data models, metadata management and advertisement services), easy-to-use tools for service development, and templates on common service patterns and types. An example of tools designed to address this need is the Introduce toolkit[15] and the caCORE Software Development Kit[16]. The Introduce toolkit facilitates the easy development and deployment of strongly-typed, secure Grid services. caGrid defines a standard set of core interfaces for caGrid compliant data services. The Introduce toolkit has a plug-in, which generates all the standard required interfaces for a caGrid data service. The caCORE SDK implements support to take an object-oriented data model, which is described in UML and registered in caDSR and annotated using controlled vocabularies in EVS, and create an object-oriented database on a relational database system. Using both tools together, a developer can go from a high level definition of a data model in Unified Modeling Language (UML) to a strongly-typed Grid data service, the backend of which is layered on top of a relational database engine, relatively easily. Nevertheless, additional tools and infrastructure support are needed in several areas to facilitate integration of resources to a service-oriented, model-driven Grid environment. We now discuss some of these areas.
There are an increasing number of applications that use XML and RDF/OWL to represent and manage datasets and semantic information. These applications will benefit from XML and RDF/OWL data services. For XML data, tools will need to be able to support mapping from XML schemas to common data elements and controlled vocabularies and create XML based backend databases. Mechanisms to translate between the common query language of caGrid and XML query languages such as XPath and XQuery. With RDF/OWL data services, an added challenge is the incorporation of semantic querying and reasoning capabilities in the environment. Moreover, extensions to the caDSR, EVS, and GME infrastructure will be needed to support publication of RDF/OWL Ontology definitions. Support is also needed to be able to easily map Grid-level object models to existing relational, XML, and RDF/OWL databases.
The Multi-scale Template, and the Coordinated Template to an extent, can involve processing of large volumes of image data, including three dimensional (time dependent) reconstruction, feature detection and annotation of 3-D microscopy imagery. This requires high performance analytical services, the backend of which should leverage distributed memory clusters, filter/stream based high performance computing, multi-core systems, SMP systems, and parallel file systems.
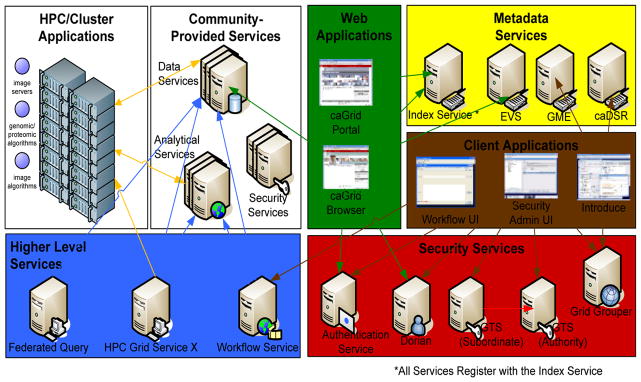
Leveraging applications on high performance architectures as self describing, interoperable, and secure services will require: 1) the design and development of gateway architectures and 2) deployment of efficient interfaces that support large scale data exchange between homogeneous or heterogeneous collections of processors. The caGrid effort is currently working on a prototype implementation of a TeraGrid Gateway service which will expose traditional HPC applications to a SOA environment. The challenge in this effort is to take a weakly typed HPC application, which utilizes the computing and data storage power of TeraGrid, and expose this application as a model driven, strongly-typed, and secure Grid service. Solving this challenge will require: Generating and registering XML schema based data models to represent the input and output data of the HPC application, mapping these syntactic data models to a community curated common semantic ontology, and generating a Grid service which will utilize these data models as input and output and can manage the execution of the HPC application. Tooling is also needed to enable HPC application authors to describe their performance tuning and job execution parameters and expose them through the Grid service. If we were to map this process to the caGrid infrastructure, the service would be like the one in Figure 2. The result of this process is that the notion the service is utilizing an HPC based application to process the request and generate a response is hidden by the Grid interface. This encapsulation will enable these applications to be seamlessly used in Grid workflows and applications without any custom standards, execution environment, and credential provisioning. The service can be discovered, semantically and syntactically, and invoked in the same fashion as any other service in the grid and still leverage the power of traditional HPC/Cluster based computing environments.
Figure 2.
A gateway service for HPC applications.
In some cases, a service may expose a data-parallel HPC application and a workflow may include multiple such services that exchange data with each other. When two parallel programs communicate with each other, distributed data structures need to be exchanged -- the data structure distributed across the nodes of a parallel program is redistributed and consumed by the nodes of the other program[17]. A decade ago, our research group developed prototype tools to support parallel data redistribution; this work has been continued and refined by Sussman.
To support such a use case efficiently, tools and infrastructure need to support 1) a distributed data descriptor (DDD) interface. A client or a service can use this interface to interrogate how data is distributed across the backend nodes of the data-parallel service[18]. 2) A parallel data transfer interface. This interface would return a handle that specifies N end-point ports, where N is the number of nodes the data-parallel service backend is executed on. The consumer of the data, which can be a parallel service itself, would be able to use the handle to exchange distributed data structures with the parallel service over an N-way channel, thus executing a parallel MxN data communication operation[18], where M is the number of nodes used by the consumer.
3.3. Federated Query and Orchestration of Grid Services
The main objective of gathering data in a scientific study is to better understand the problem being studied and to be able to predict, explain, and extrapolate potential solutions to the problem and possible outcomes. This process requires complex problem solving environments that integrate modeling of the analysis process, on demand access, and processing of heterogeneous and very large databases. Compelling workflow requirements arise from all three Design Templates described here. Studies in the Coordinated Template, for instance, involve querying and assimilating information associated with multiple groups of subjects, comparing and correlating the information about the subject under study with this information, and classifying the analysis results. Data obtained from queries into various databases available in the environment are processed through a series of simple and complex operations including subsetting, correction of various data acquisition artifacts, filtering operations, feature detection, feature classification, as well as interactive inspection and annotation of data.
The caGrid infrastructure provides support for federated querying of multiple data services to enable distributed aggregation and joins on object classes and object associations defined in domain object models. The current support for federated query is aimed at the basic functionality required for data subsetting and integration. Extensions to this basic support are needed to provide more comprehensive support for the design templates.
Scalability of federated query support is important when there are large numbers of clients and queries span large volumes of data and a large number of services. Middleware components need to be developed that will enable distributed execution of queries by using HPC systems available in the environment as well as by carefully creating sub-queries, pushing them to services or groups of services for execution, and coordinating data exchange between services to minimize communication overheads. Several projects have investigated and developed mechanisms for query execution in distributed environments [19–30]. A suite of coordination services can be developed to implement techniques developed in those projects.
Another architecture requirement for federated query components is the support for semantic queries. Semantic federated query support is particularly important for the Coordinated Template and the Multiscale Template, where datasets and features extracted in these datasets can be annotated with concepts and properties from one or more ontologies, creating a domain-specific knowledge base. Middleware components are needed to support queries that involve selection and join criteria based not only on a data model, which represents the structure and attributes of objects in a dataset, but also on semantic annotations. A key requirement is to be able to support reasoning on ontologies based on description logic (DL) so that a richer set of queries can be executed and answered based on annotations inferred from explicit annotations. Semantic querying capabilities, reasoning techniques and tools, and runtime systems have been researched and developed in several projects [31–36]. A suite of coordination services can be developed to support semantic querying on methods developed in those projects. However, more comprehensive support for federated query of semantic information sources will require a closer integration of SOA, MDA, and Semantic Grid technologies [37–40].
caGrid also provides a workflow management service that supports the execution and monitoring of workflows expressed in the Business Process Execution Language (BPEL)[41]. One drawback of the current workflow support is that all data items transferred between services in the workflow have to be routed through the workflow management service, a bottleneck for workflows that process large volumes of data. This is a problem especially in the multiscale and coordinated templates in which large volumes of image, ECG, and microarray data may need to be exchanged between services in a workflow. When extending the workflow support, it is important to facilitate composition of Grid services into workflows without requiring any modifications to the application-specific service code; that is, the implementation of an application service (an analytical or a data service) need not depend on whether or not the service may be part of a workflow. To support this requirement, a workflow helper service, which coordinates with the workflow management service, will be needed. The helper service is directly responsible for the integration of an application service into a workflow. The helper service needs to take into consideration all the security issues involved in invoking an application service. It handles the process of receiving data from upstream services in the workflow, invoking the methods of the application service as specified in the workflow description, and staging the results from service invocations to downstream services. With the helper service, the need to route data and messages through the workflow management service is eliminated. The helper service can be deployed remotely and interacts with the application service like a client. It can also be deployed locally in the same container as the application service, saving overheads due to SOAP messages.
The helper service should be designed to be modular and extensible so that additional functionality can be added. For instance, the workflow management service has a global view of the workflow environment and is responsible for managing multiple workflows and interpreting BPEL-based workflow descriptions. The basic functionality of the helper service is to receive a method invocation command from the workflow manager service and handle method invocation and routing of input and output data. This functionality could be augmented to improve performance and enable hierarchical workflows. A workflow described in the Grid environment can involve services running on high-performance Grid nodes.
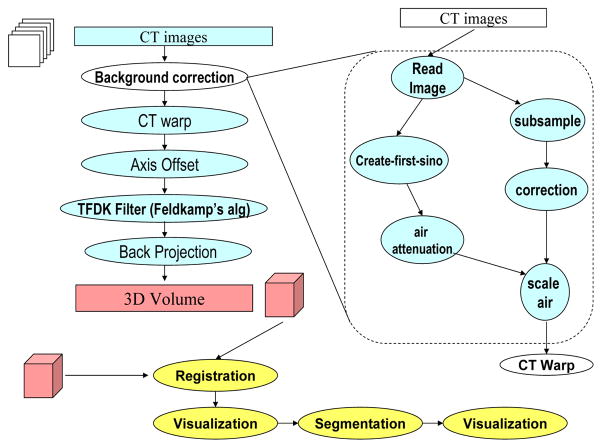
Figure 3 illustrates an example image analysis workflow for CT images. In this example, the background correction, CT wrap, axis offset, TFDK filter, and back projection operations can each be a Grid service in the environment. The background correction service consists of another series of operations. Each of these operations can be exposed as Grid service methods. They can also be formed into a workflow within the background correction service. If the background service runs on a high-performance Grid node (i.e., a node hosted on a cluster system), it can benefit from execution of its portion of the Grid workflow as a fine-grained dataflow process. In that case, a Grid workflow containing the background correction service not only includes the network of services constituting the workflow and interactions between them, but should also include the fine-grained dataflow within the service.
Figure 3.
A sample image analysis for micro-CT images. A series of methods are applied on the CT images to reconstruct a 3D volume. The background projection method in this example consists of another series of operations on data. The result 3D volume can then be registered with a 3D volume from another imaging session, registered volumes can be visualized and segmented, and segmentation results can be visualized by the researcher.
One of the challenges is to be able to express such hierarchical workflows and coordinate execution of the service level interactions and fine-grained dataflow operations within a service in the Grid level workflow. The helper service will need to be extended to handle not only simple method invocation instructions from the workflow manager service, but also more complex instructions expressing the dataflow within the service. A variety of middleware tools have been developed by our group and others to optimize execution of data analysis workflows as dataflow networks on heterogeneous compute and storage clusters[24, 28, 42, 43]. The helper service will allow integration of these middleware tools to enable execution of dataflow networks within services. With such extensions, workflow scheduling now becomes a hierarchical process, and should take into consideration, for optimizing both the coarse grain (i.e., Grid-level workflow) and the fine grain (i.e., dataflow within the service) components.
Considering HPC applications implemented from high level languages, compiler support for hierarchical workflow frameworks are needed. For such compilers, the functional decomposition of the applications plays an important role. Moreover, Grid environments are dynamic, meaning that the final decomposition of application processing into workflow components should be decided at runtime, and there should be room for adapting the generated code as the conditions change during execution. We have referred to this process as telescopic compilation, in the sense that the granularity of the decomposition can be adjusted at runtime. We intend to pursue such compilation problems in our future research efforts as well.
3.4. Interoperability with Existing Institutional Systems and Other Middleware
The Prospective Template motivates the requirement for interoperability of a middleware system with existing institutional systems and other middleware. As is described in Section 2, the process of managing clinical trials and data collection requires interaction with a range of open and commercial systems. Data in these systems are represented, exchanged, and accessed through a set of architectures and standards, such as HL7, IHE, DICOM, caBIG™ Silver and Gold level, developed by different communities.
Middleware systems developed on top of a particular standard and framework will not be able to readily interact with middleware systems developed on top of other standards. caGrid, for example, is built upon the Grid Services standards. Each data and analytical resource in caGrid is implemented as a Grid Service, which interacts with other resources and clients using Grid Service protocols. caGrid services are standard Web Service Resource Framework (WSRF v1.2)[4] services and can be accessed by any specification-compliant client. Although WSRF makes use of XML technologies for data representation and exchange, it is not directly compatible with HL7 and IHE, which also use XML. A set of tools and services are needed to enable efficient mappings between different messaging standards, controlled vocabularies, and data types associated with many communities. Some tools will be used to harmonize an external standard for data representation with the common data models and ontologies accepted by a community so that semantic interoperability with external systems can be achieved. Other tools and services will be employed as gateways between different protocols to support on-the-fly transformation of messages and resource invocations.
3.5. Security
Protection of sensitive data and intellectual property is an important component in many design templates. The Prospective Template in particular has strong requirements for authentication and controlled access to data because of the fact that prospective clinical research studies capture, reference, and manage patient related information. While security concerns are less stringent in the other design templates, protection of intellectual property and proprietary resources is important. In biomedical domain, there are several institutional, state-wide, and federal regulations on who can access sensitive data and how such data can be accessed and should be protected. The Grid Authentication and Authorization with Reliably Distributed Services (GAARDS) infrastructure of caGrid provides a comprehensive set of services and tools for the administration and enforcement of security policy in an enterprise Grid[44]. Nevertheless, best practices, policy, and infrastructure are required to meet the increasing demands of Grid environments. These additional components are needed to address requirements associated with compliance with federal e-authentication guidelines, compliance with regulatory policy, establishment of a Grid-wide user directory.
Compliance with Federal e-Authentication Guidelines
The caBIG program has chosen to adopt the Federal e-Authentication initiative (http://www.cio.gov/eauthentication/), which provides guidelines for authentication. The guidelines describe four levels of assurance, levels one through four, each with increasingly restrictive guidelines for authentication. With each increasing level service providers have increased assurance of the identity of the client they are communicating with. The caBIG community has adopted GAARDS Dorian as their account management system. Dorian issues a Grid account to users based on their existing accounts provided by their organization. The level assurance of a Grid account is determined based on minimum of the following: The level of assurance that the GAARDS Dorian infrastructure can obtain and the level of assurance of the participating organization with the lowest level of assurance. Currently, Dorian can facilitate management of accounts for level one and level two assurances. In general the GAARDS and to a larger extent the Grid infrastructure is already capable of supporting level 3 and level 4. In the future additional tools or extensions to existing tools such as Dorian will need to be developed for managing and provisioning level 3 and level 4 accounts.
Individual organizations vary in terms of the levels of assurance that they can currently meet. Our current research indicates that the majority of organizations affiliated with the caBIG community are operating at about a level 1. Bringing the organizations to level 2, level 3, and level 4 present major challenges and will require the development of best practices, statement of procedures, and tools to aid them in doing so. A scalable framework for evaluating and auditing organizations for compliance with the e-authentication guidelines will also be required.
Compliance with Regulatory Guidelines
Ensuring that the Grid infrastructure meets regulatory guidelines such as 21CRF Part 11 and HIPAA is critical in being able to exchange protected health information (PHI). Beginning with Dorian, the caGrid infrastructure is undergoing a review for compliance with regulatory guidelines. To meet regulatory guidelines additional infrastructure will need to be developed to allow service providers to meet the auditing requirements. Furthermore additional documentation, best practices, statement of procedures, and policies will need to be developed.
Establishment of a Grid-wide User Directory
In the caBIG community authorization and access control requirements vary significantly amongst service providers, because of this the caBIG community has chosen to leave authorization up to individual service providers. GAARDS provides tools that enable service providers to make authorization decisions. These tools include GAARDS Grid Grouper and the caCORE Common Security Module (CSM). Both Grid Grouper and CSM enforce access control policy based on a client’s Grid identity. Although this solution is very effective it becomes difficult to provision access control policy because in order to allow a client access to a resource you need their Grid identity. In a large, federated environment it can be difficult to confidently determine one’s Grid identity. For example, if one wanted to allow John Doe to access to a resource, they would need to know John Doe’s Grid identity. However, there are some challenges. How do we confidently determine John Doe’s Grid identity and what if there is more than one John Doe? Once John Doe’s identity has been determined, how confident are we that it is correct? To help alleviate these difficulties in the future and to support other similar use cases a Grid-wide user directory is required containing accurate information about users. If such a directory were to exist and service providers were confident in the accuracy of its information, then this directory could be used in conjunction with tools like Grid Grouper and CSM to more easily provision access control policy.
4. Conclusions
Service oriented (SOA) and model driven (MDA) architectures have tremendous potential to facilitate more effective and efficient utilization of disparate data and analytical resources and address requirements arising from common design templates in scientific research. The caGrid system is a realization of the merging of the MDA and SOA paradigms with an emphasis on common data elements and controlled vocabularies. While it provides a comprehensive suite of core services and tools, there is still room for improvement in several areas. In this paper we have discussed requirements that arise in design templates in the translational research domain. We described ideas on architecture features in middleware systems to address these requirements. We focused on several core areas including support for data and analytical services, semantic information management, federated query and workflows, integration of HPC applications, and security. We believe that additional research and development in these and other areas such as interoperability between systems building on standards developed by different communities will further promote and facilitate a wider use of MDA and SOA technologies in science, biomedicine, and engineering.
Acknowledgments
This work was supported in part by the National Cancer Institute (NCI) caGrid Developer grant 79077CBS10; the State of Ohio Board of Regents BRTT Program grants ODOD AGMT TECH 04-049 and BRTT02-0003; funds from The Ohio State University Comprehensive Cancer Center-Arthur G. James Cancer Hospital and Richard J. Solove Research Institute; the National Science Foundation (NSF) grants: CNS-0615155, CNS-0403342, CCF-0342615, CNS-0426241, and CNS-0406386; the NHLBI R24 HL085343 grant; and the National Institutes of Health (NIH) U54 CA113001 and R01 LM009239 grants.
Bibliography
- 1.Alexander C. A Pattern Language: Towns, Buildings, Construction (Center for Environmental Structure Series) Oxford University Press; USA: 1977. [Google Scholar]
- 2.Gamma E, Helm R, Johnson R, Vlissides JM. Design Patterns: Elements of Reusable Object-Oriented Software (Addison-Wesley Professional Computing Series) Addison-Wesley Professional; 1994. [Google Scholar]
- 3.Graham S, Simeonov S, Boubez T, Davis D, Daniels G, Nakamura Y, Neyama R. Building Web Services with Java: Making Sense of XML, SOAP, WSDL, and UDDI. SAMS Publishing; 2002. [Google Scholar]
- 4.Foster I, Czajkowski K, Ferguson D, Frey J, Graham S, Maguire T, Snelling D, Tuecke S. Modeling and Managing State in Distributed Systems: The Role of OGSI and WSRF. Proceedings of IEEE. 2005;93:604–612. [Google Scholar]
- 5.Oster S, Hastings S, Langella S, Ervin D, Madduri R, Kurc T, Siebenlist F, Foster I, Shanbhag K, Covitz P, Saltz J. caGrid 1.0: A Grid Enterprise Architecture for Cancer Research. Proceedings of the 2007 American Medical Informatics Association (AMIA) Annual Symposium; Chicago, IL. 2007. [PMC free article] [PubMed] [Google Scholar]
- 6.Saltz J, Oster S, Hastings S, Kurc T, Sanchez W, Kher M, Manisundaram A, Shanbhag K, Covitz P. caGrid: Design and Implementation of the Core Architecture of the Cancer Biomedical Informatics Grid. Bioinformatics. 2006;22:1910–1916. doi: 10.1093/bioinformatics/btl272. [DOI] [PubMed] [Google Scholar]
- 7.caBIG Compatibility Guidelines. 2005. https://cabig.nci.nih.gov/guidelines_documentation/caBIGCompatGuideRev2_final.pdf.
- 8.Fridsma DB, Evans J, Hastak S, Mead CN. The BRIDG Project: A Technical Report. Journal of American Medical Informatics Association (JAMIA) 2008 doi: 10.1197/jamia.M2556. PrePrint: Accepted Article. Published December 20, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Quaranta WAV, Cummings PT, Anderson AR. Mathematical modeling of cancer: the future of prognosis and treatment. Clin Chim Acta. 2005;357:173–179. doi: 10.1016/j.cccn.2005.03.023. [DOI] [PubMed] [Google Scholar]
- 10.Oster S, Langella S, Hastings S, Ervin D, Madduri R, Phillips J, Kurc T, Siebenlist F, Covitz P, Shanbhag K, Foster I, Saltz J. caGrid 1.0: An Enterprise Grid Infrastructure for Biomedical Research. Journal of American Medical Informatics Association (JAMIA) 2008 doi: 10.1197/jamia.M2522. PrePrint: Accepted Article. Published December 20, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Covitz PA, Hartel F, Schaefer C, Coronado S, Fragoso G, Sahni H, Gustafson S, Buetow KH. caCORE: A Common Infrastructure for Cancer Informatics. Bioinformatics. 2003;19:2404–2412. doi: 10.1093/bioinformatics/btg335. [DOI] [PubMed] [Google Scholar]
- 12.Hastings S, Langella S, Oster S, Saltz J. Distributed Data Management and Integration: The Mobius Project. Proceedings of the Global Grid Forum 11 (GGF11) Semantic Grid Applications Workshop; Honolulu, Hawaii, USA. 2004. pp. 20–38. [Google Scholar]
- 13.Allcock B, Bester J, Bresnahan J, Chervenak A, Foster I, Kesselman C, Meder S, Nefedova V, Quesnal D, TS Data Management and Transfer in High Performance Computational Grid Environments. Parallel Computing Journal. 2002;28:749–771. [Google Scholar]
- 14.Allcock B, Breshanan J, Kettimuthu R, Link M, Dumitrescu C, Raicu I, Foster I. The Globus Striped GridFTP Framework and Server. Supercomputing 2005 (SC 2005) 2005 [Google Scholar]
- 15.Hastings S, Oster S, Langella S, Ervin D, Kurc T, Saltz J. Introduce: An Open Source Toolkit for Rapid Development of Strongly Typed Grid Services. Journal of Grid Computing. 2007;5:407–427. [Google Scholar]
- 16.Phillips J, Chilukuri R, Fragoso G, Warzel D, Covitz PA. The caCORE Software Development Kit: Streamlining construction of interoperable biomedical information services. BMC Medical Informatics and Decision Making. 2006;6 doi: 10.1186/1472-6947-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee J-Y, Sussman A. High performance communication between parallel programs. Proceedings of 2005 Joint Workshop on High-Performance Grid Computing and High-Level Parallel Programming Models (HIPS-HPGC 2005); IEEE Computer Society Press; 2005. [Google Scholar]
- 18.Wu JS, Sussman A. Flexible control of data transfers between parallel programs. Proceedings of the Fifth International Workshop on Grid Computing - GRID 2004; IEEE Computer Society Press; 2004. pp. 226–234. [Google Scholar]
- 19.Andrade H, Kurc T, Sussman A, Saltz J. Active Proxy-G: Optimizing the Query Execution Process in the Grid. Proceedings of the ACM/IEEE Supercomputing Conference (SC2002); Baltimore, MD: ACM Press/IEEE Computer Society Press; 2002. [Google Scholar]
- 20.Bell WH, Bosio D, Hoschek W, Kunszt P, McCance G, Silander M. Project Spitfire -- Towards Grid Web Service Databases. Global Grid Forum Informational Document, GGF5; Edinburgh, Scotland. 2002. [Google Scholar]
- 21.Beynon M, Kurc T, Catalyurek U, Chang C, Sussman A, Saltz J. Distributed Processing of Very Large Datasets with DataCutter. Parallel Computing. 2001;27:1457–2478. [Google Scholar]
- 22.Chang C, Kurc T, Sussman A, Catalyurek U, Saltz J. A Hypergraph-Based Workload Partitioning Strategy for Parallel Data Aggregation. Proceedings of the Tenth SIAM Conference on Parallel Processing for Scientific Computing; Portsmouth, VA. 2001. [Google Scholar]
- 23.DeWitt D, Gray J. Parallel Database Systems: the Future of High Performance Database Systems. Communications of the ACM. 1992;35:85–98. [Google Scholar]
- 24.Isert C, Schwan K. ACDS: Adapting Computational Data Streams for High Performance. 14th International Parallel & Distributed Processing Symposium (IPDPS 2000); Cancun, Mexico: IEEE Computer Society Press; 2000. [Google Scholar]
- 25.Kossman D. The State of the Art in Distributed Query Processing. ACM Computing Surveys. 2000;32:422–469. [Google Scholar]
- 26.Narayanan S, Catalyurek U, Kurc T, Saltz J. Applying Database Support for Large Scale Data Driven Science in Distributed Environments. Proceedings of the 4th International Workshop on Grid Computing (Grid 2003); 2003. [Google Scholar]
- 27.Narayanan S, Kurc T, Catalyurek U, Saltz J. Database Support for Data-driven Scientific Applications in the Grid. Parallel Processing Letters. 2003;13:245–273. [Google Scholar]
- 28.Plale B, Schwan K. dQUOB: Managing Large Data Flows Using Dynamic Embedded Queries. IEEE International High Performance Distributed Computing (HPDC); 2000. [Google Scholar]
- 29.Sheth AP, Larson JA. Federated Database Systems for Managing Distributed, Heterogeneous, and Autonomous Databases. ACM Computing Surveys. 1990;22:183–236. [Google Scholar]
- 30.Smith J, Gounaris A, Watson P, Paton NW, Fernandes AA, Sakellariou R. Distributed Query Processing on the Grid. Proceedings of the Third Workshop on Grid Computing (GRID2002); Baltimore, MD. 2002. [Google Scholar]
- 31.Broekstra J, Kampman A, van Harmelen F. Sesame: A generic architecture for storing and querying RDF and RDF schema. International Semantic Web Conference, Lecture Notes in Computer Science; 2002. pp. 54–68. [Google Scholar]
- 32.Harris S, Gibbins N. 3store: Efficient bulk RDF storage. 1st International Workshop on Practical and Scalable Semantic Web Systems held at ISWC} 2003, volume 89 of CEUR Workshop Proceedings. CEUR-WS. org; 2003. [Google Scholar]
- 33.Wilkinson K, Sayers C, Kuno HA, Reynolds D. Efficient RDF storage and retrieval in Jena2. Proceedings of VLDB Workshop on Semantic Web and Databases; 2003. pp. 131–150. [Google Scholar]
- 34.Haarslev V, Moller R. Racer: A core inference engine for the semantic web. 2nd International Workshop on Evaluation of Ontology-based Tools (EON 2003); 2003. [Google Scholar]
- 35.Horrocks I. The FaCT system. Proceedings of the International Conference on Automated Reasoning with Analytic Tableaux and Related Methods (TABLEAUX 98), volume 1397 of LNAI; 1998. pp. 307–312. [Google Scholar]
- 36.Kiryakov A, Ognyanov D, Manov D. OWLIM - A pragmatic semantic repository for OWL. WISE Workshops, volume 3807 of Lecture Notes in Computer Science; 2005. pp. 182–192. [Google Scholar]
- 37.Semantic Grid Community Portal. 2008. http://www.semanticgrid.org/
- 38.Corcho O, Alper P, Kotsiopoulos I, Missier P, Bechhofer S, Goble CA. An overview of S-OGSA: A Reference Semantic Grid Architecture. Journal of Web Semantics. 2006;4:102–115. [Google Scholar]
- 39.Goble CA, Kesselman C, Sure Y. Semantic Grid: The Convergence of Technologies. Internationales Begegnungs- und Forschungszentrum für Informatik (IBFI); 2005. [Google Scholar]
- 40.Newhouse S, Mayer A, Furmento N, McGough S, Stanton J, Darlington J. Laying the Foundations for the Semantic Grid. Proceedings of the AISB’02 Symposium on AI and GRID Computing; 2002. [Google Scholar]
- 41.Business Process Execution Language for Web Services, Version 1.0. 2002. http://www-106.ibm.com/developerworks/webservices/library/ws-bpel/
- 42.Beynon M, Chang C, Catalyurek U, Kurc T, Sussman A, Andrade H, Ferreira R, Saltz J. Processing Large-Scale Multidimensional Data in Parallel and Distributed Environments. Parallel Computing. 2002;28:827–859. [Google Scholar]
- 43.Ferreira RA, Meira W, Guedes D, Drummond LMA, Coutinho B, Teodoro G, Tavares T, Araujo R, Ferreira GT. Anthill: a scalable run-time environment for data mining applications. 17th International Symposium on Computer Architecture and High Performance Computing (SBAC-PAD 2005); 2005. [Google Scholar]
- 44.Langella S, Oster S, Hastings S, Siebenlist F, Phillips J, Ervin D, Permar J, Kurc T, Saltz J. The Cancer Biomedical Informatics Grid (caBIG™) Security Infrastructure. Proceedings of the 2007 American Medical Informatics Association (AMIA) Annual Symposium; Chicago, IL. 2007. [PMC free article] [PubMed] [Google Scholar]