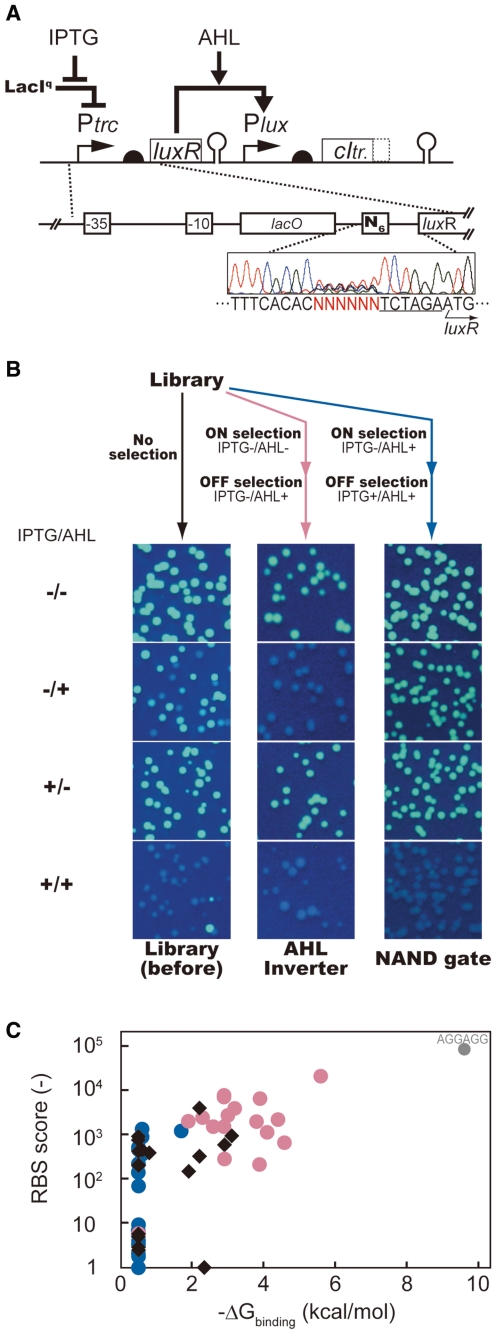
Figure 7.
Isolating AHL Inverters and NAND-gates from a single rbs library. (A) Library design. The six bases (N6 in figure) corresponding to rbs of luxR gene were randomized. (B) The phenotype variation of the circuit library before (left), after the ‘AHL-inverter selection’ (middle), and after the ‘NAND-gate selection’ (right). (C) Distribution of the rbs scores in the variants from original (black filled diamonds), AHL inverter-selected (red filled circles), and NAND gate-selected (blue filled circles) libraries. X-axis represents the free binding energy of these rbs sequences (ΔGbinding) and anti-rbs sequence, calculated using the RNA duplex software component of the Vienna RNA Package (45). Rbs scores for Y-axis were calculated using RBS Calculator (14). The sequence AGGAGG (perfect match to the anti-rbs) was independently constructed by site-directed mutagenesis. It was found to behave as an AHL inverter.