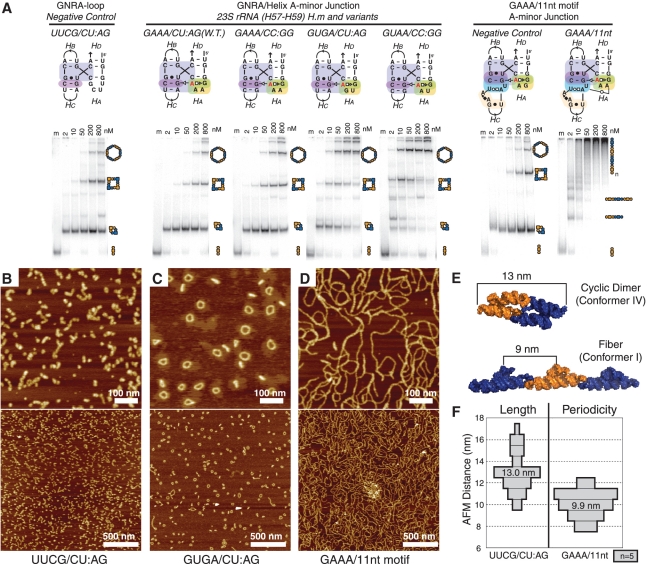
Figure 3.
Analysis of different A-minor 4WJ motifs. (A) Native PAGE at 1 mM Mg(OAc)2 and TB 1× showing the end-to-end assembly of tectoRNAs containing different GNRA/helix and GNRA/receptor A-minor junction sequence signatures. Each experiment includes one lane (lane m) loaded with only one molecule (A) of the two (A+B), as a monomer standard. The other lanes in each set contain equal concentrations (nanomolar) of the two molecules A+B, assembled at 30°C in presence of 1 mM Mg(OAc)2, 50 mM KCl and TB 1×buffer. Higher molecular weight products are indicative of increased stability of conformer I, while low molecular weight products likely represent assemblies forced into conformer IV by the entropic closure of KL interactions. (B–D) A 600-nm scale AFM images were acquired in air under tapping mode. RNA samples were prepared at 200 nM, in 1 mM Mg(OAc)2, 50 mM KCl, TB 1× buffer and were adjusted to 100 nM RNA and 8 mM Mg(OAc)2 immediately before deposition on mica and imaging. (B) Complexes formed from UUCG/CU:AG A-minor junction motif. (C) Complexes formed from GUGA/CU:AG A-minor junction motifs. (D) Complexes formed from the GAAA/11-nt A-minor junction hybrid motif. (E) 3D computer model showing the expected size of the cyclic dimer and the expected periodicity of bulky features on the fiber (see also Supplementary Figure S5). (F) Histogram showing the average measured size of cyclic dimers observed in the UUCG 4WJ sample and average measured periodicity measured for fibers in the GAAA/11-nt-motif sample. W.T., wild type; H.m, H. marismortui.